Search Count: 13
 |
Design And Application Of Synthetic 17B-Hsd13 Substrates To Drug Discovery, And To Reveal Preserved Catalytic Activity Of Protective Human Variants
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2024-11-27 Classification: HYDROLASE Ligands: NAD, A1AG4 |
 |
Design And Application Of Synthetic 17B-Hsd13 Substrates To Drug Discovery, And To Reveal Preserved Catalytic Activity Of Protective Human Variants
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2024-11-27 Classification: HYDROLASE Ligands: NAD, A1AG5 |
 |
Design And Application Of Synthetic 17B-Hsd13 Substrates To Drug Discovery, And To Reveal Preserved Catalytic Activity Of Protective Human Variants
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2024-11-27 Classification: HYDROLASE Ligands: NAD, A1AG6 |
 |
Jnk3 (Mitogen-Activated Protein Kinase 10) In Complex With Compound 23 Bearing A C(Sp3)F2Br Moiety
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2023-08-02 Classification: TRANSFERASE Ligands: SWM, EDO, GOL, BME, PEG, C15, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-11-09 Classification: TRANSFERASE Ligands: IWU, PG4, PEG, PGE, GOL, EPE, 1PE, SO4, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2022-11-09 Classification: CELL CYCLE Ligands: LE9, EPE, ZN, PEG |
 |
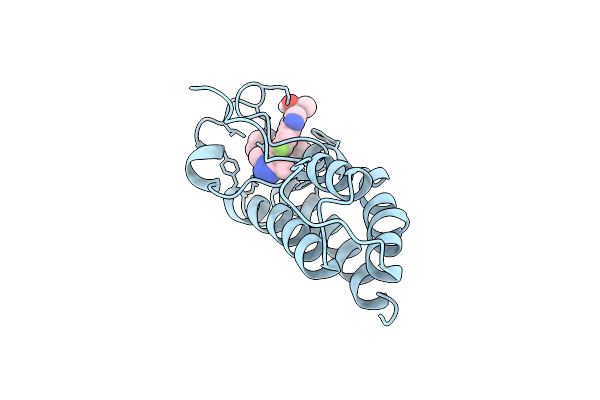
Crystal Structure Of The First Bromodomain Of Human Brd4 In Complex With Bmt-206059 Aka 2-{(3M)-3-(1,4-Dimethyl-1H-1,2,3-Triazol-5-Yl)-8-Fluoro-5-[(S)-(Oxan-4-Yl)(Phenyl)Methyl]-5H-Pyrido[3,2-B]Indol-7-Yl}Propan-2-Ol, Triply Deuterated On The 4-Methyl Group
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2022-08-17 Classification: CELL CYCLE Ligands: PQF, NA, EDO |
 |
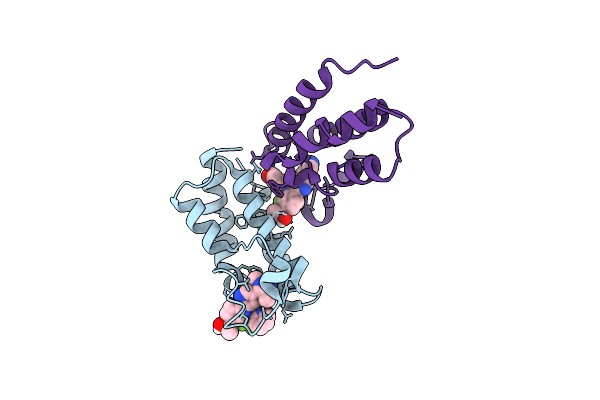
Crystal Structure Of The First Bromodomain Of Human Brd4 In Complex With 2-{(7P)-7-(1,4-Dimethyl-1H-1,2,3-Triazol-5-Yl)-8-Fluoro-5-[(S)-(Oxan-4-Yl)(Phenyl)Methyl]-5H-Pyrido[3,2-B]Indol-3-Yl}Propan-2-Ol
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2021-09-29 Classification: CELL CYCLE Ligands: YWY |
 |
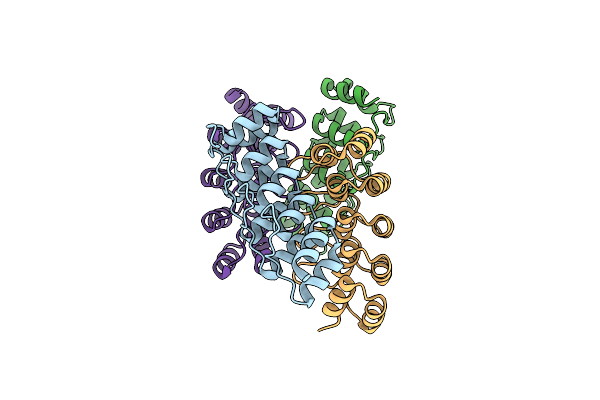
Crystal Structure Of The First Bromodomain Of Human Brd4 In Complex With 2-{3-(1,4-Dimethyl-1H-1,2,3-Triazol-5-Yl)-6-Fluoro-5-[(S)-(3-Fluoropyridin-2-Yl)(Oxan-4-Yl)Methyl]-5H-Pyrido[3,2-B]Indol-7-Yl}Propan-2-Ol
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2021-05-26 Classification: CELL CYCLE Ligands: YX4 |
 |
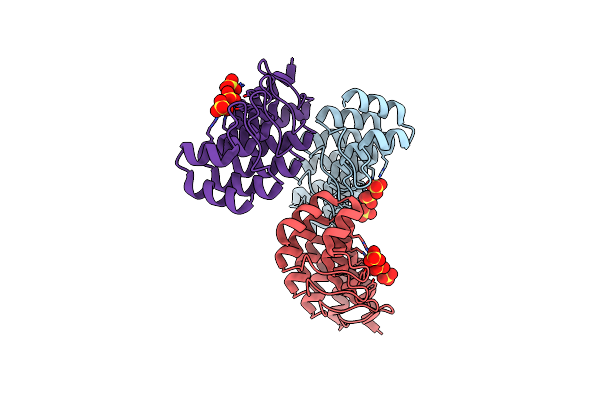
Structural Determinants For Improved Thermal Stability Of Designed Ankyrin Repeat Proteins With A Redesigned C-Capping Module.
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-08-18 Classification: DE NOVO PROTEIN |
 |
Structural Determinants For Improved Thermal Stability Of Designed Ankyrin Repeat Proteins With A Redesigned C-Capping Module.
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2010-08-18 Classification: DE NOVO PROTEIN Ligands: SO4 |
 |
Structural Determinants For Improved Thermal Stability Of Designed Ankyrin Repeat Proteins With A Redesigned C-Capping Module.
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-08-18 Classification: DE NOVO PROTEIN Ligands: EDO, MOH, SO4 |
 |
Crystal Structure Of Plasmodium Falciparum Dihydroorotate Dehydrogenase Bound With Novel Inhibitor Genz667348
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-08-18 Classification: OXIDOREDUCTASE Ligands: O8A, FMN, ORO, LDA |

