Search Count: 16
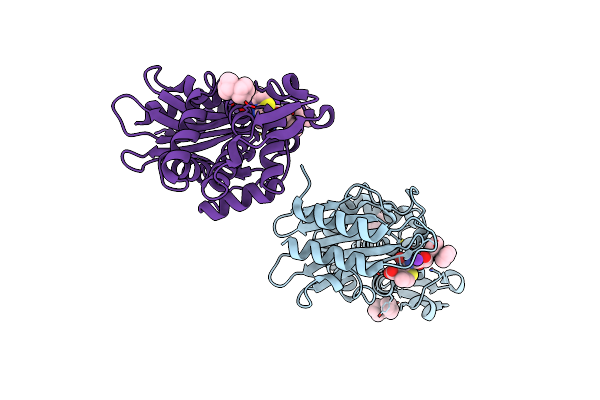 |
X-Ray Crystal Structure Of Ruthenocenyl-7-Aminocephalosporanic Acid Covalent Acyl-Enzyme Complex With Ctx-M-14 E166A Beta-Lactamase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2020-04-01 Classification: HYDROLASE Ligands: 7G4, TDJ, K |
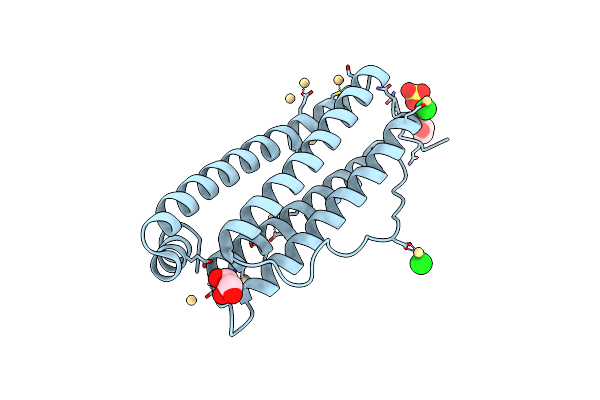 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2019-02-06 Classification: METAL TRANSPORT Ligands: CD, CL, SO4, GOL |
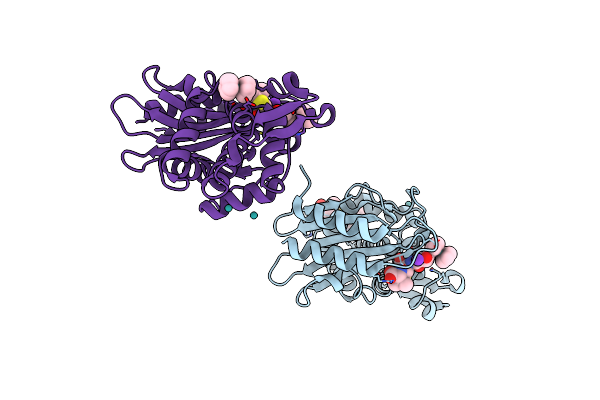 |
Atomic Resolution X-Ray Crystal Structure Of A Ruthenocene Conjugated Beta-Lactam Antibiotic In Complex With Ctx-M-14 S70G Beta-Lactamase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2017-11-08 Classification: HYDROLASE/ANTIBIOTIC Ligands: K, JSE, RU, JSC, 7G4 |
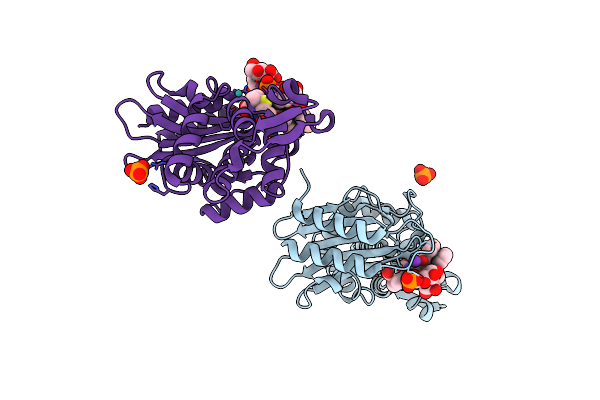 |
X-Ray Crystal Structure Of Ruthenocene Conjugated Penicilloate And Penilloate Products In Complex With Ctx-M-14 E166A Beta-Lactamase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2017-11-08 Classification: HYDROLASE/ANTIBIOTIC Ligands: K, PO4, JSC, JSD, RU |
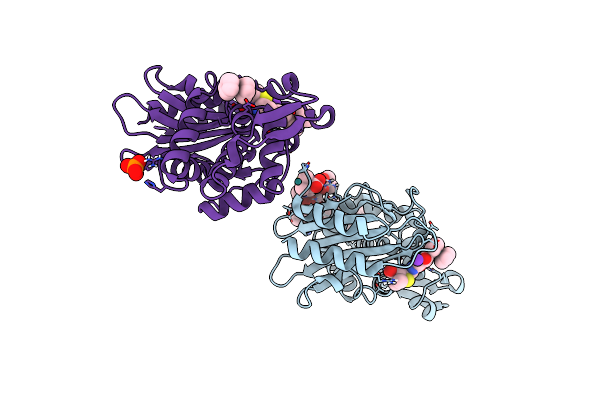 |
Ultrahigh Resolution X-Ray Crystal Structure Of Ruthenocene Conjugated Penicilloate And Penilloate Products In Complex With Ctx-M-14 E166A Beta-Lactamase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:0.85 Å Release Date: 2017-11-08 Classification: HYDROLASE Ligands: JSD, JSC, JSE, K, RU, PO4 |
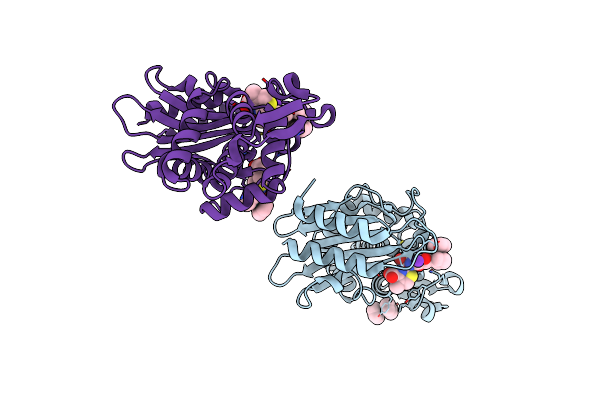 |
X-Ray Crystal Structure Of Ruthenocenyl-7-Aminodesacetoxycephalosporanic Acid Covalent Acyl-Enyzme Complex With Ctx-M-14 E166A Beta-Lactamase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2017-02-08 Classification: HYDROLASE Ligands: K, 8CY, LSI |
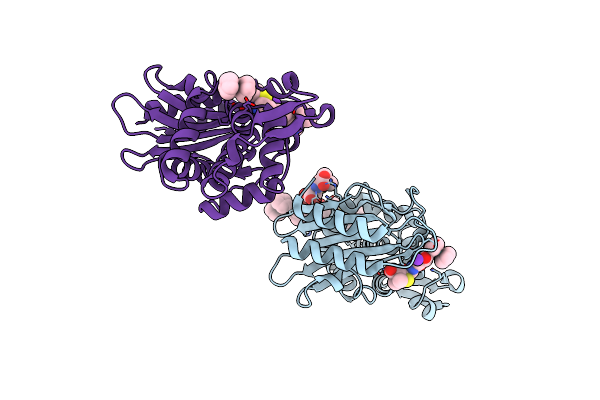 |
Atomic Resolution X-Ray Crystal Structure Of A Ruthenocene Conjugated Beta-Lactam Antibiotic In Complex With Ctx-M-14 E166A Beta-Lactamase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2015-03-18 Classification: HYDROLASE/ANTIBIOTIC Ligands: K, JSD, JSC, JSE |
 |
 |
 |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2005-08-09 Classification: PROTEIN BINDING |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2005-08-09 Classification: OXIDOREDUCTASE, APOPTOSIS |
 |
Solution Structure Of A Cchh Mutant Of The Ninth Cchc Zinc Finger Of U-Shaped
Organism: Drosophila melanogaster
Method: SOLUTION NMR Release Date: 2002-09-25 Classification: TRANSCRIPTION Ligands: ZN |
 |
Solution Structure Of The Ninth Zinc-Finger Domain Of The U-Shaped Transcription Factor
Organism: Drosophila melanogaster
Method: SOLUTION NMR Release Date: 2000-10-04 Classification: TRANSCRIPTION Ligands: ZN |
 |
Solution Structure Of The First Zinc Finger From The Drosophila U-Shaped Transcription Factor
Organism: Drosophila melanogaster
Method: SOLUTION NMR Release Date: 2000-10-04 Classification: TRANSCRIPTION Ligands: ZN |
 |
Solution Structure Of The N-Terminal Zinc Finger Of Murine Gata-1, Nmr, 25 Structures
Organism: Mus musculus
Method: SOLUTION NMR Release Date: 1999-06-08 Classification: TRANSCRIPTION REGULATION Ligands: ZN |

