Search Count: 7
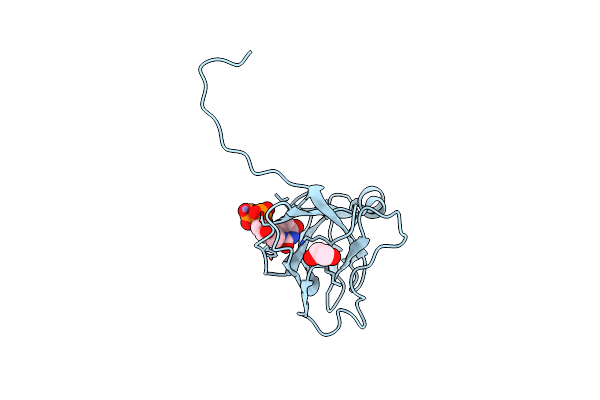 |
Crystal Structure Of Dutpase In Complex With Substrate Analogue Dudp And Manganese
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2007-07-31 Classification: HYDROLASE Ligands: MN, DUD, EDO |
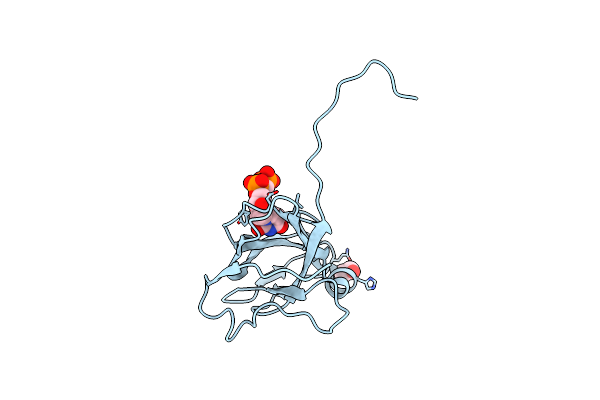 |
Crystal Structure Of Dutpase Complexed With Substrate Analogue Methylene-Dutp
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2007-07-31 Classification: HYDROLASE Ligands: UC5, EDO |
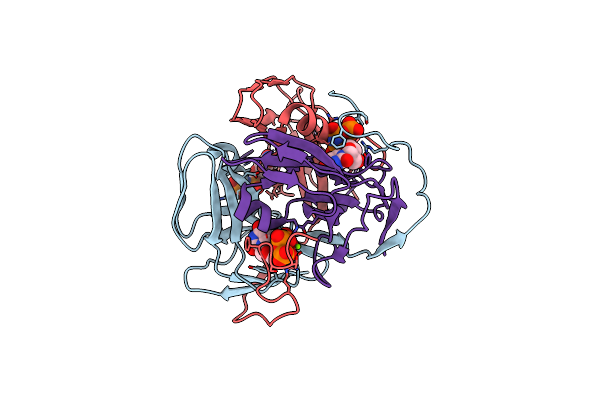 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-07-24 Classification: HYDROLASE Ligands: MG, DUP, CL |
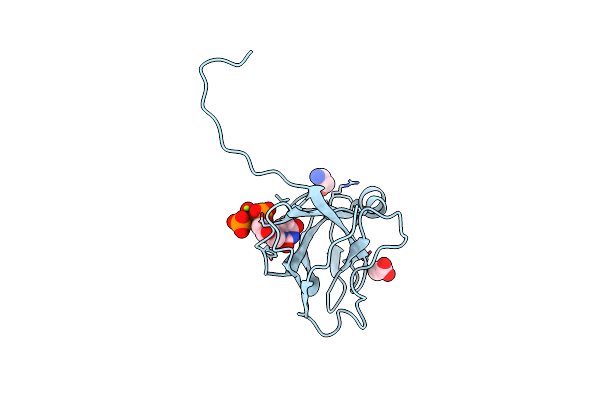 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2004-09-07 Classification: HYDROLASE Ligands: MG, ACT, DUP, TRS |
 |
Crystal Structure Of Inactive Mutant Dutpase Complexed With Substrate Analogue Imido-Dutp
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2004-09-07 Classification: HYDROLASE Ligands: MG, DUP, TRS |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2004-09-07 Classification: HYDROLASE Ligands: UMP, TRS |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2004-09-07 Classification: HYDROLASE Ligands: MG, DUT, TRS |

