Search Count: 91
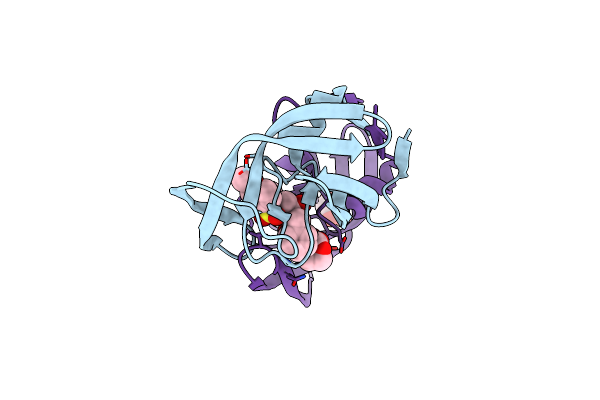 |
Crystal Structure Of A Highly Resistant Hiv-1 Protease Clinical Isolate Pr10X With Grl-0519 (Tris-Tetrahydrofuran As P2 Ligand)
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2022-10-05 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: G52, CL, GOL |
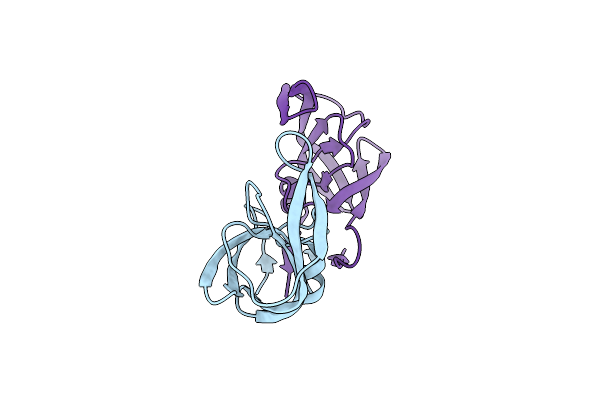 |
Crystal Structure Of A Highly Resistant Hiv-1 Protease Clinical Isolate Pr10X (Inhibitor-Free)
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2022-10-05 Classification: VIRAL PROTEIN, HYDROLASE |
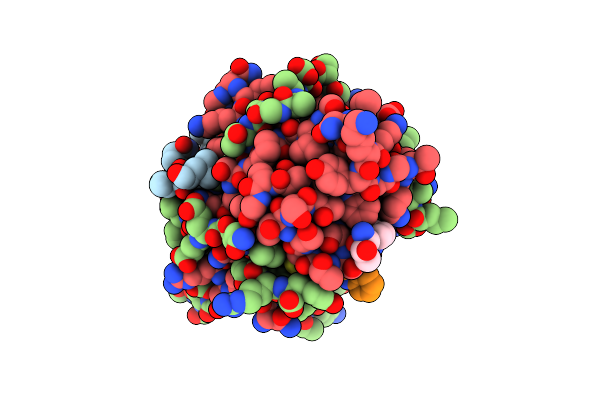 |
Crystal Structure Of Deuterated Gamma-Chymotrypsin At Ph 7.5, Room Temperature
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: IOD |
 |
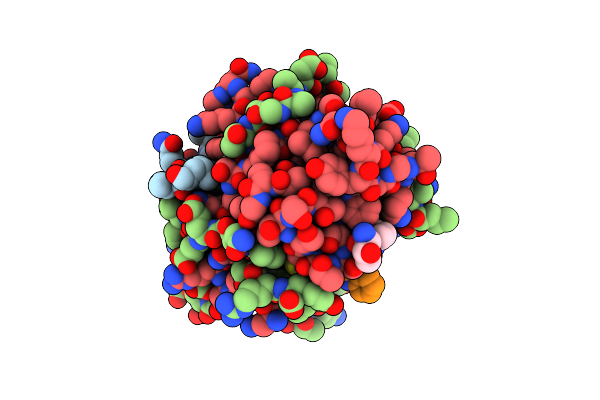
Crystal Structure Of Deuterated Gamma-Chymotrypsin At Ph 7.5, Cryo Temperature
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: MLA, IOD |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: IOD |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: MLI, IOD |
 |
Crystal Structure Of Deuterated Gamma-Chymotrypsin At Ph 5.6, Room Temperature
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: IOD, SO4 |
 |
Crystal Structure Of Deuterated Gamma-Chymotrypsin At Ph 5.6, Cryo Temperature
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: MLA, IOD |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: IOD, SO4 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: MLI, IOD |
 |
Crystal Structure Of Deuterated Gamma-Chymotrypsin At Ph 9, Room Temperature
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: IOD, SO4 |
 |
Crystal Structure Of Deuterated Gamma-Chymotrypsin At Ph 9, Cryo Temperature
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: SO4, IOD |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: IOD, SO4 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: IOD, SO4 |
 |
Room Temperature X-Ray Crystallographic Structure Of A Jonesia Denitrificans Lytic Polysaccharide Monooxygenase At 1.1 Angstrom Resolution.
Organism: Jonesia denitrificans (strain atcc 14870 / dsm 20603 / cip 55134)
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2017-05-24 Classification: SUGAR BINDING PROTEIN Ligands: CU, PER |
 |
Neutron Crystallographic Structure Of A Jonesia Denitrificans Lytic Polysaccharide Monooxygenase
Organism: Jonesia denitrificans (strain atcc 14870 / dsm 20603 / cip 55134)
Method: NEUTRON DIFFRACTION Resolution:2.10 Å Release Date: 2017-05-24 Classification: SUGAR BINDING PROTEIN Ligands: CU, PER |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-03-01 Classification: TRANSFERASE |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2017-03-01 Classification: TRANSFERASE |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2017-03-01 Classification: TRANSFERASE |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-03-01 Classification: TRANSFERASE |

