Search Count: 145
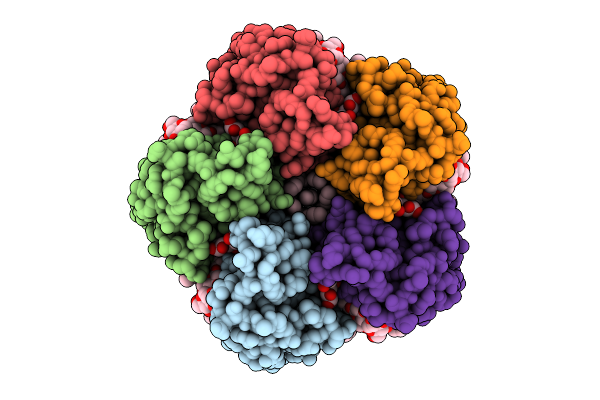 |
Cryo-Em Structure Of The Light-Driven Sodium Pump Ernar In The Pentameric Form
Organism: Erythrobacter
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: LFA, LMT, RET |
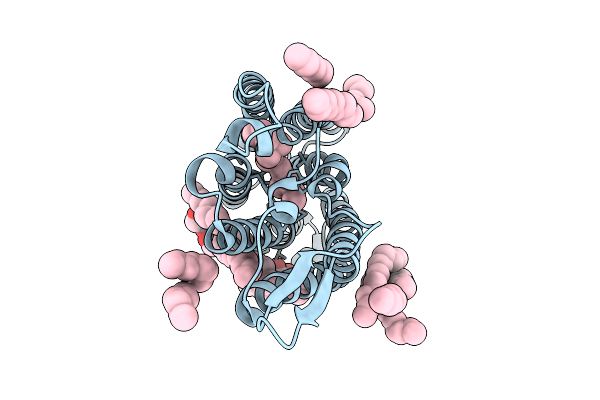 |
Cryo-Em Structure Of The Light-Driven Sodium Pump Ernar In The Monomeric Form In The K2 State
Organism: Erythrobacter
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: LFA, LMT, RET |
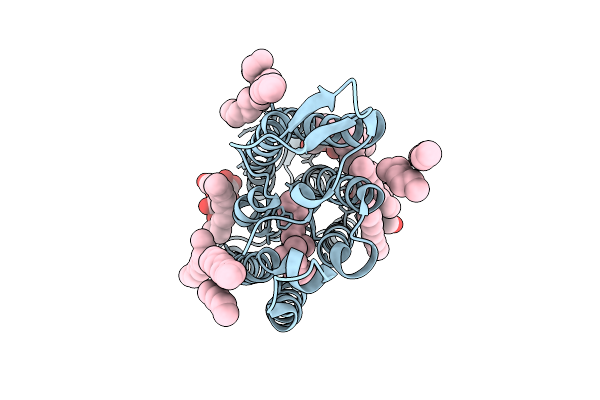 |
Cryo-Em Structure Of The Light-Driven Sodium Pump Ernar In The Monomeric Form In The O2 State
Organism: Erythrobacter
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: LFA, LMT, RET |
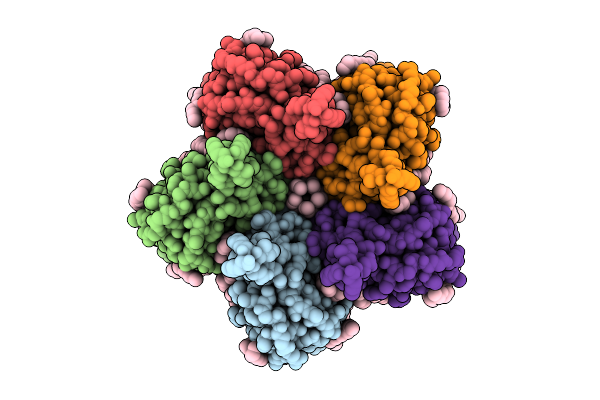 |
Cryo-Em Structure Of The Flotillin-Associated Rhodopsin Psfar In Detergent Micelle
Organism: Candidatus pseudothioglobus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: LFA |
 |
Cryo-Em Structure Of The Light-Driven Proton Pump Pspr In Detergent Micelle
Organism: Candidatus pseudothioglobus sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: LFA, RET, LMT |
 |
Cryo-Em Structure Of The Double Mutant H84V/E120G Of The Flotillin-Associated Rhodopsin Psfar In Detergent Micelle
Organism: Candidatus pseudothioglobus sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
Structure Of Meiothermus Ruber Mrub_1259 Lov Domain With N- And C-Terminal Alpha Helices (Mrlove)
Organism: Meiothermus ruber dsm 1279
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: FLUORESCENT PROTEIN Ligands: FMN |
 |
Organism: Meiothermus ruber dsm 1279
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: FLUORESCENT PROTEIN Ligands: FMN |
 |
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LMT, LFA, RET |
 |
Cryo-Em Structure Of The Microbial Rhodopsin Cryor1 At Ph 10.5 In Detergent In The Ground State
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LMT, RET, LFA |
 |
Cryo-Em Structure Of The Microbial Rhodopsin Cryor1 At Ph 10.5 In Detergent In The M State
Organism: Cryobacterium levicorallinum
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LMT, RET |
 |
Organism: Subtercola endophyticus
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The Ground State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, GOL, RET, PO4 |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The P596 State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, GOL, RET, PO4 |
 |
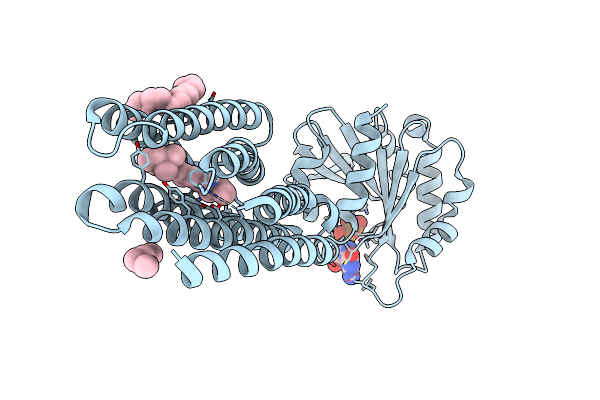
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) - Human Gtpase Arf1 (L8K,Q71L) Chimera; Ground State
Organism: Candidatus actinomarina minuta, Homo sapiens, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: GDP, LFA, RET |
 |
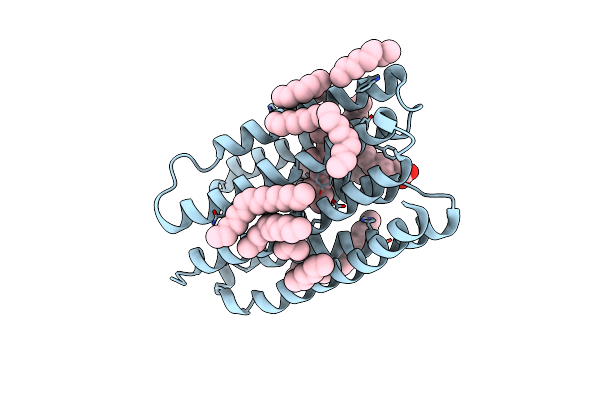
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) - Human Gtpase Arf1 (L8K,Q71L) Chimera; N State
Organism: Candidatus actinomarina minuta, Homo sapiens, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: GDP, LFA, OLA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O* State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O* State, Ph 8.8
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |

