Search Count: 22
 |
Crystal Structure Bovine Hsc70(Aa1-554)E213A/D214A In Complex With Nicotinic-Acid-Derivative
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2022-09-07 Classification: CHAPERONE Ligands: 7UE, K, EPE, GOL |
 |
Crystal Structure Of Bovine Hsc70(Aa1-554)E213A/D214A In Complex With Triazine-Derivative
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2022-06-01 Classification: CHAPERONE Ligands: EPE, K, GOL, V8Q |
 |
Crystal Structure Of Bovine Hsc70(Aa1-554)E213A/D214A In Complex With Tricine
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2022-06-01 Classification: CHAPERONE Ligands: GOL, VLS, EPE, K |
 |
Crystal Structure Of Bovine Hsc70(Aa1-554)E213A/D214A In Complex With Methanesulfonamide
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2022-06-01 Classification: CHAPERONE Ligands: EPE, V7Z, GOL, NA |
 |
Crystal Structure Of Bovine Hsc70(Aa1-554)E213A/D214A In Complex With 1H-Indazole
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-05-04 Classification: CHAPERONE Ligands: EPE, LZ1, DMS, GOL, K, TMO |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2021-12-15 Classification: DNA-RNA HYBRID |
 |
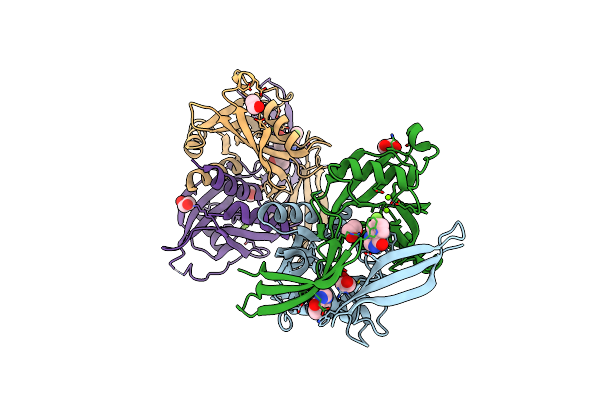
Pandda Analysis Group Deposition -- Crystal Structure Of Nudt5 In Complex With Gb-0804
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: MG, CL, EDO, PW7 |
 |
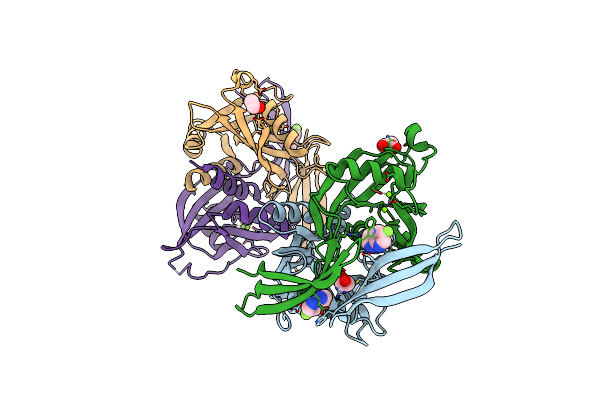
Pandda Analysis Group Deposition -- Crystal Structure Of Nudt5 In Complex With Fs-2639
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: MG, CL, PWA, EDO |
 |
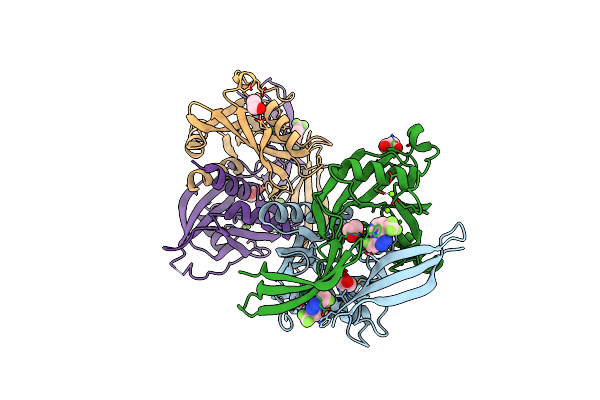
Pandda Analysis Group Deposition -- Crystal Structure Of Nudt5 In Complex With Ss-4432
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: MG, CL, PWD, EDO |
 |
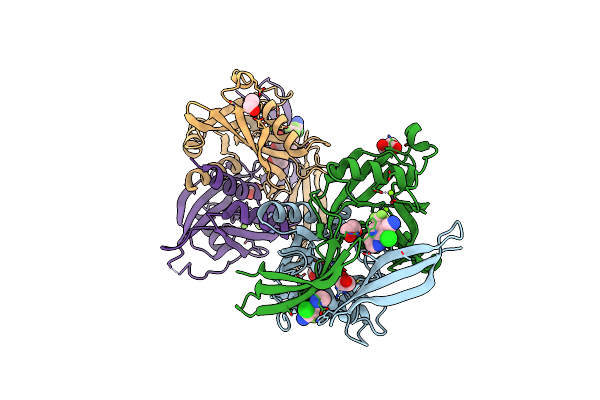
Pandda Analysis Group Deposition -- Crystal Structure Of Nudt5 In Complex With 1R-0641
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: MG, CL, PWG, EDO |
 |
Pandda Analysis Group Deposition -- Crystal Structure Of Nudt5 In Complex With Ae-0227
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: MG, PWJ, EDO, CL |
 |
Pandda Analysis Group Deposition -- Crystal Structure Of Nudt5 In Complex With Fs-1169
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: MG, PWM, EDO, CL |
 |
Pandda Analysis Group Deposition -- Crystal Structure Of Nudt5 In Complex With Fs-3764
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: MG, EDO, PWP |
 |
Pandda Analysis Group Deposition -- Crystal Structure Of Nudt5 In Complex With 8J-537S
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: MG, CL, PWS, EDO |
 |
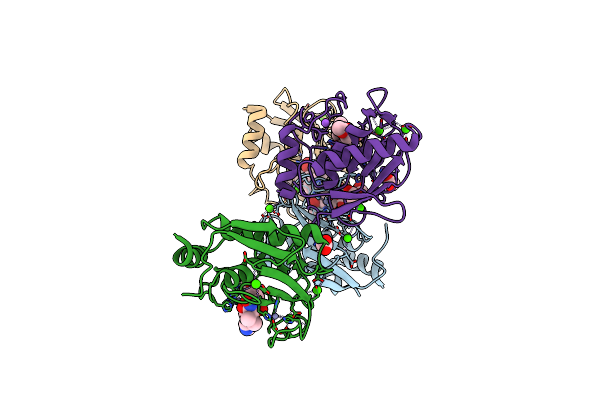
Crystal Structure Of Bovine Hsc70(Aa1-554)E213A/D214A In Complex With Inhibitor Ver155008
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2019-08-14 Classification: CHAPERONE Ligands: TMO, 3FD, EPE, GOL |
 |
Does A Fast Nuclear Magnetic Resonance Spectroscopy- And X-Ray Crystallography Hybrid Approach Provide Reliable Structural Information Of Ligand-Protein Complexes? A Case Study Of Metalloproteinases.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-03-03 Classification: HYDROLASE Ligands: ZN, CA, ACT, CGS, CL, NA |
 |
Organism: Echis carinatus
Method: SOLUTION NMR Release Date: 2003-12-09 Classification: CELL ADHESION |
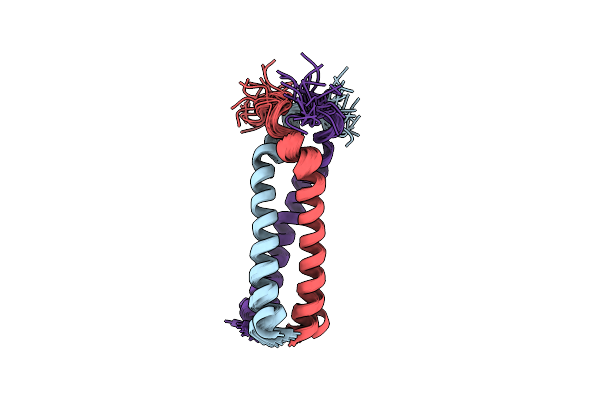 |
Solution Structure Of The Coiled-Coil Trimerization Domain From Lung Surfactant Protein D
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2002-11-27 Classification: SUGAR BINDING PROTEIN |
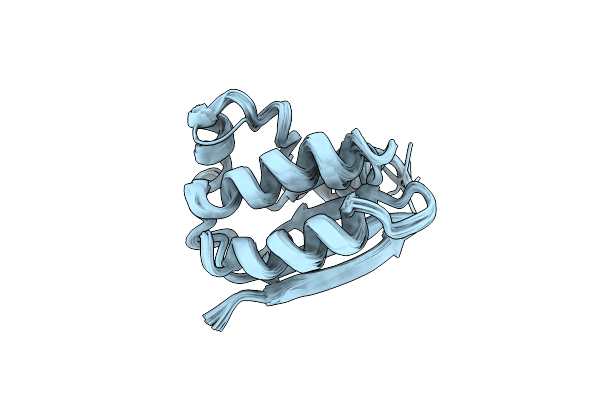 |
Solution Structure Of Spoiiaa, A Phosphorylatable Component Of The System That Regulates Transcription Factor Sigma-F Of Bacillus Subtilis, Nmr, 24 Structures
Organism: Bacillus subtilis
Method: SOLUTION NMR Release Date: 1998-07-01 Classification: TRANSCRIPTION REGULATOR |
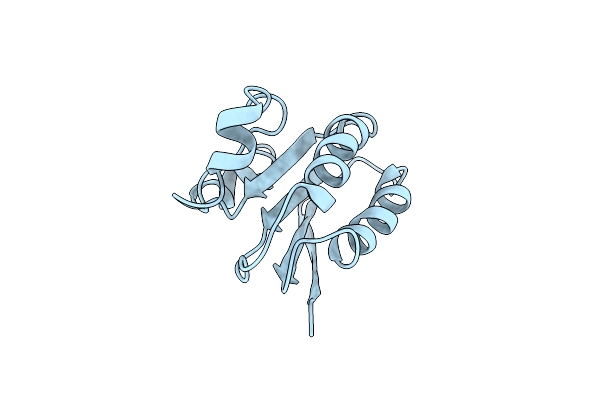 |
Solution Structure Of Spoiiaa, A Phosphorylatable Component Of The System That Regulates Transcription Factor Sigma-F Of Bacillus Subtilis Nmr, Minimized Average Structure
Organism: Bacillus subtilis
Method: SOLUTION NMR Release Date: 1998-07-01 Classification: TRANSCRIPTION REGULATOR |

