Search Count: 84
 |
Structure Of The C-Terminal Protease Ctpa-Lbca Complex Of Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: HYDROLASE |
 |
The C-Terminal Protease Ctpa-Lbca Complex Of Pseudomonas Aeruginosa With The Tpr At The High Position
Organism: Pseudomonas aeruginosa, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: HYDROLASE |
 |
The C-Terminal Protease Ctpa-Lbca Complex Of Pseudomonas Aeruginosa With The Tpr At The Low Position
Organism: Pseudomonas aeruginosa, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: HYDROLASE |
 |
Structure Of The C-Terminal Protease Ctpa-Lbca Complex Of Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: HYDROLASE |
 |
Structure Of The Mycobacterium Tuberculosis Hsp70 Protein Dnak Bound To The Nucleotide Exchange Factor Grpe
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: CHAPERONE |
 |
Structure Of The Mycobacterium Tuberculosis 20S Proteasome Bound To The C-Terminal Gqyl Motif Of The Atp-Bound Mpa Atpase
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2022-08-03 Classification: ANTIMICROBIAL PROTEIN |
 |
Structure Of The Mycobacterium Tuberculosis 20S Proteasome Bound To The C-Terminal Gqyl Motif Of The Adp-Bound Mpa Atpase
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2022-08-03 Classification: ANTIMICROBIAL PROTEIN |
 |
Structure Of The Mycobacterium Tuberculosis 20S Proteasome Bound To The Atp-Bound Mpa Atpase
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2022-08-03 Classification: ANTIMICROBIAL PROTEIN |
 |
Structure Of The Mycobacterium Tuberculosis 20S Proteasome Bound To The Adp-Bound Mpa Atpase
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2022-08-03 Classification: ANTIMICROBIAL PROTEIN |
 |
Crystal Structure Of Carboxyl-Terminal Processing Protease A, Ctpa, Of Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2022-04-27 Classification: HYDROLASE |
 |
Crystal Structure Of Lbca (Lipoprotein Binding Partner Of Ctpa) Of Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2022-04-27 Classification: PROTEIN BINDING |
 |
Crystal Structure Of Carboxyl-Terminal Processing Protease A Mutant S302A, Ctpa_S302A, Of Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2022-04-27 Classification: HYDROLASE |
 |
Cryo-Em Structure Of E. Coli P Pilus Tip Assembly Intermediate Papc-Papd-Papk-Papg In The First Conformation
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-08-11 Classification: CHAPERONE |
 |
Cryo-Em Structure Of E. Coli P Pilus Tip Assembly Intermediate Papc-Papd-Papk-Papg In The Second Conformation
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-08-11 Classification: CHAPERONE |
 |
Cryo-Em Structure Of E. Coli P Pilus Tip Assembly Intermediate Papc-Papd-Papk-Papf-Papg
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-08-11 Classification: CHAPERONE |
 |
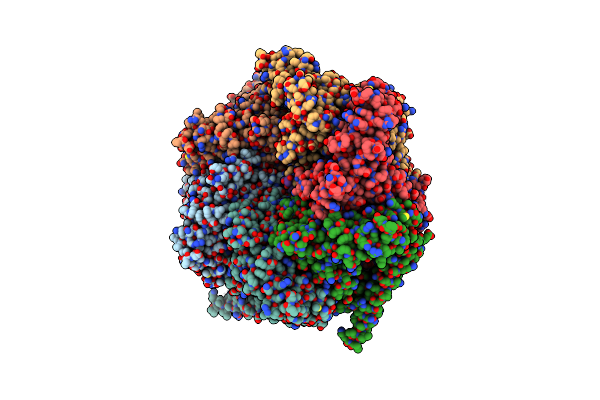
The Mycobacterium Tuberculosis Clpb Disaggregase Hexamer Structure With A Locally Refined Clpb Middle Domain And A Dnak Nucleotide Binding Domain
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2021-05-26 Classification: CHAPERONE Ligands: AGS, ADP |
 |
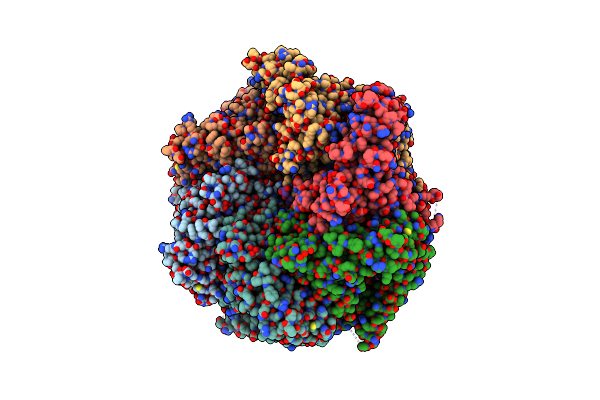
The Mycobacterium Tuberculosis Clpb Disaggregase Hexamer Structure In Conformation Ii In The Presence Of Dnak Chaperone And A Model Substrate
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2021-05-26 Classification: CHAPERONE Ligands: AGS, ADP |
 |
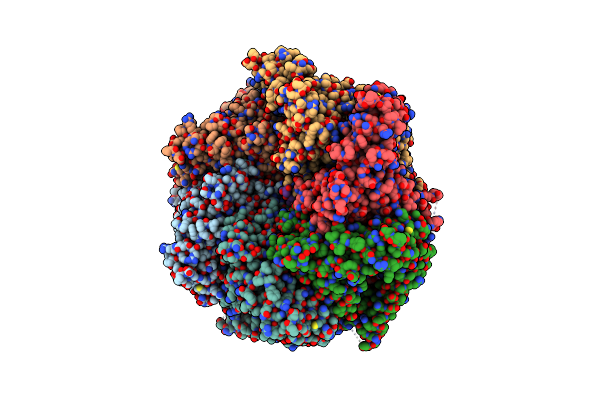
The Mycobacterium Tuberculosis Clpb Disaggregase Hexamer Structure In Conformation T In The Presence Of Dnak Chaperone And A Model Substrate
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2021-05-26 Classification: CHAPERONE Ligands: AGS, ADP |
 |
The Mycobacterium Tuberculosis Clpb Disaggregase Hexamer Structure With A Locally Refined N-Terminal Domain In The Presence Of Dnak Chaperone And A Model Substrate
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2021-05-26 Classification: CHAPERONE Ligands: AGS, ADP |
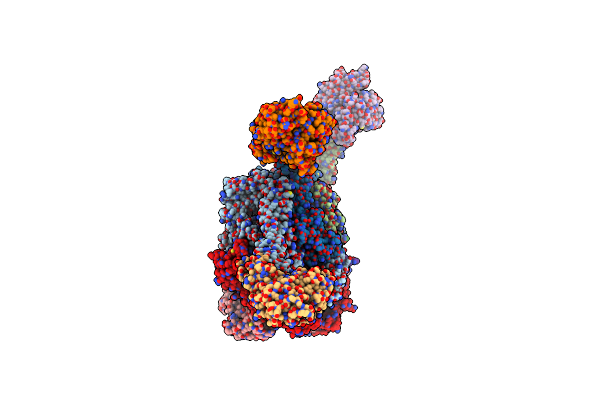 |
The Mycobacterium Tuberculosis Clpb Disaggregase Hexamer Structure With Three Locally Refined Clpb Middle Domains And Three Dnak Nucleotide Binding Domains
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2021-05-26 Classification: CHAPERONE Ligands: AGS, ADP |

