Search Count: 30
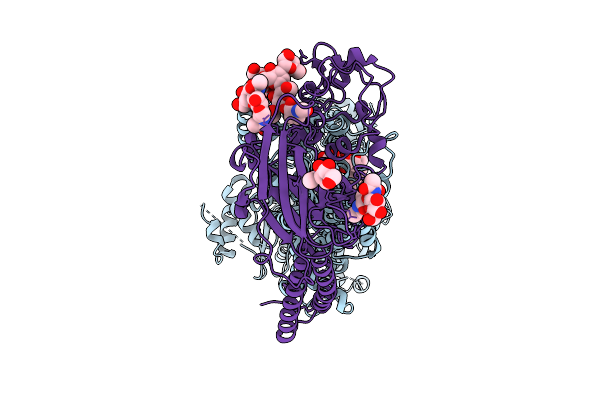 |
Cryo-Em Structure Of The C. Neoformans Lipid Flippase Apt1-Cdc50 Bound With Butyrolactol A In The E2P State
Organism: Cryptococcus neoformans var. grubii h99
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: TRANSLOCASE/INHIBITOR Ligands: NAG, BEF, MG, A1BD6 |
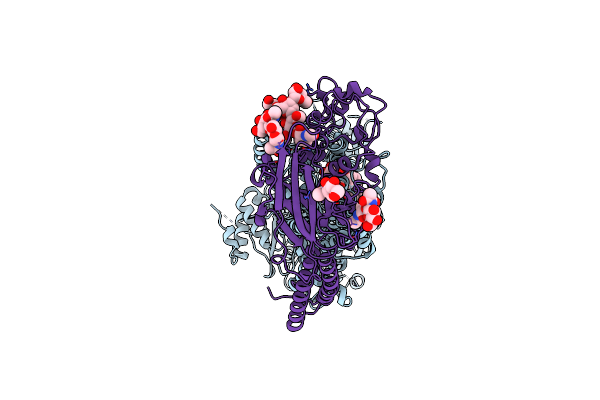 |
Cryo-Em Structure Of The C. Neoformans Lipid Flippase Apt1-Cdc50 Bound With Butyrolactol A In The E2P State
Organism: Cryptococcus neoformans var. grubii h99
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: TRANSLOCASE/INHIBITOR Ligands: NAG, BEF, MG, A1BD6 |
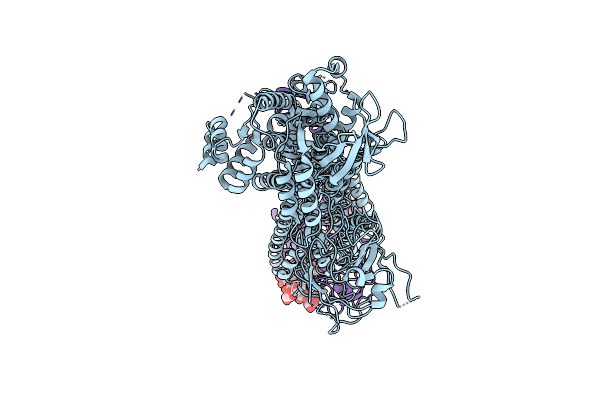 |
Cryo-Em Structure Of The C. Neoformans Lipid Flippase Apt1-Cdc50 In The E1 State
Organism: Cryptococcus neoformans var. grubii h99
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: LIPID TRANSPORT Ligands: NAG, MG |
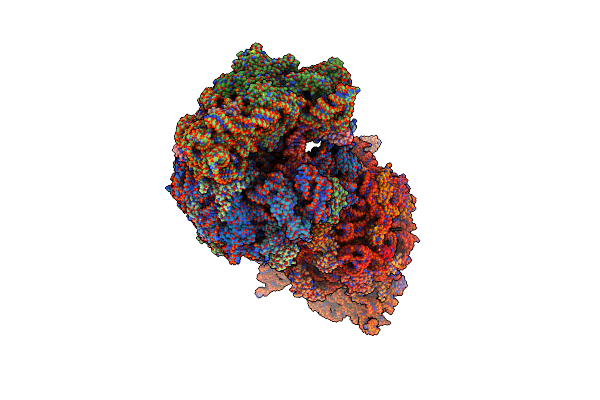 |
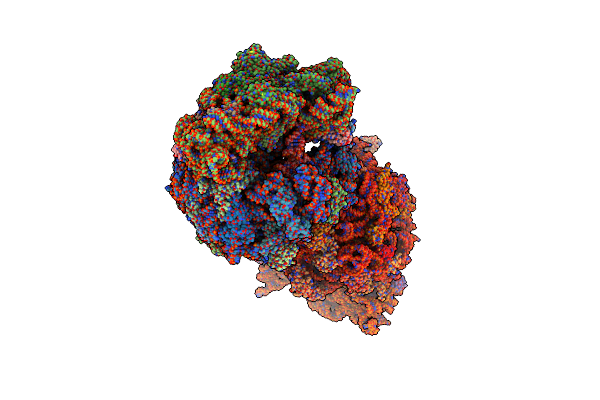
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Lasso Peptide Lariocidin, Mrna, Aminoacylated A-Site Phe-Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.50A Resolution
Organism: Escherichia coli, Paenibacillus, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG, K, ZN, SF4 |
 |
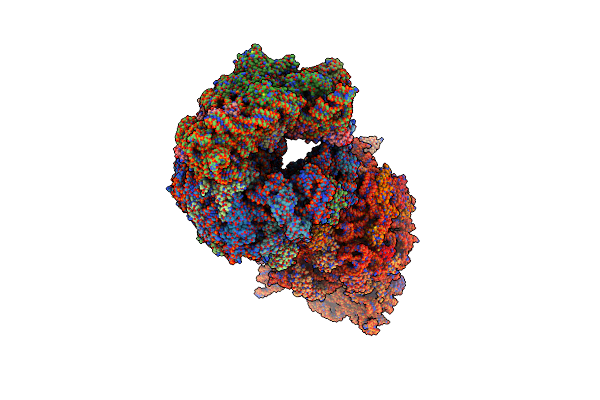
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Lasso Peptide Lariocidin B, Mrna, Aminoacylated A-Site Phe-Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.60A Resolution
Organism: Escherichia coli, Paenibacillus, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG, K, ZN, SF4 |
 |
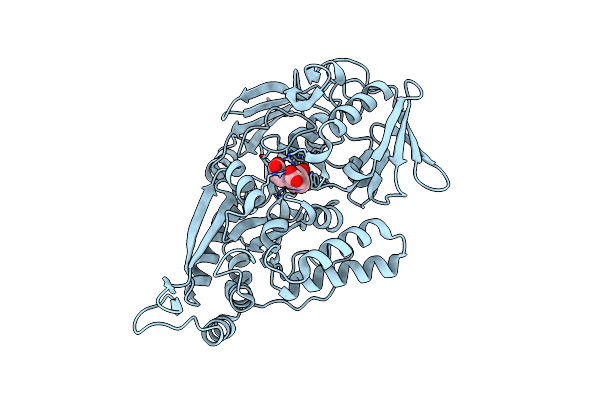
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Lasso Peptide Lariocidin And Protein Y At 2.60A Resolution
Organism: Escherichia coli k-12, Paenibacillus, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG, ARG, MPD, ZN, SF4 |
 |
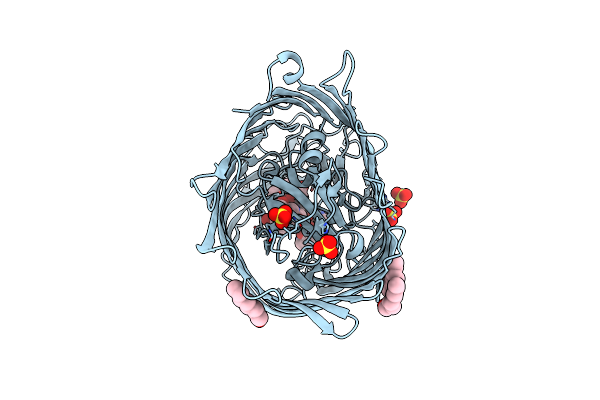
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-04-10 Classification: TRANSPORT PROTEIN Ligands: II1, NI |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2023-04-26 Classification: MEMBRANE PROTEIN Ligands: SO4, BOG, FE, OX8 |
 |
Crystal Structure Of Aminoglycoside Resistance Enzyme Apma, Complex With Gentamicin
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2023-04-19 Classification: TRANSFERASE Ligands: LLL, EDO, MG, CL |
 |
Crystal Structure Of Aminoglycoside Resistance Enzyme Apma, Complex With Tobramycin
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2023-04-19 Classification: TRANSFERASE Ligands: TOY, CL |
 |
Crystal Structure Of Aminoglycoside Resistance Enzyme Apma, Complex With Kanamycin B And Coenzyme A
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2022-11-02 Classification: TRANSFERASE Ligands: 9CS, COA, EDO, SO4, CL |
 |
Crystal Structure Of Aminoglycoside Resistance Enzyme Apma, Complex With Paromomycin And Coenzyme A
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2022-11-02 Classification: TRANSFERASE Ligands: COA, PAR, GOL |
 |
Crystal Structure Of Aminoglycoside Resistance Enzyme Apma, Complex With Neomycin
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2022-11-02 Classification: TRANSFERASE Ligands: NMY, EDO |
 |
Crystal Structure Of Aminoglycoside Resistance Enzyme Apma H135A Mutant, Complex With Tobramycin And Coenzyme A
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2022-11-02 Classification: TRANSFERASE Ligands: TOY, EDO, COA |
 |
Organism: Enterococcus casseliflavus
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2018-02-28 Classification: TRANSFERASE/ANTIBIOTIC Ligands: CL, EDS |
 |
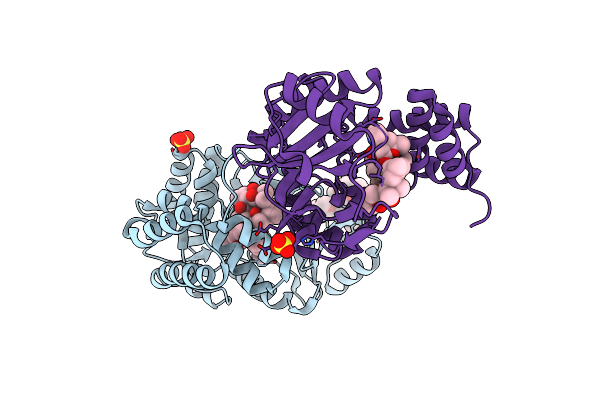
Crystal Structure Of Rifampin Monooxygenase From Streptomyces Venezuelae, Complexed With Rifampin And Fad
Organism: Streptomyces venezuelae (strain atcc 10712 / cbs 650.69 / dsm 40230 / jcm 4526 / nbrc 13096 / pd 04745)
Method: X-RAY DIFFRACTION Resolution:3.32 Å Release Date: 2017-12-13 Classification: OXIDOREDUCTASE Ligands: FAD, RFP, CL, MG |
 |
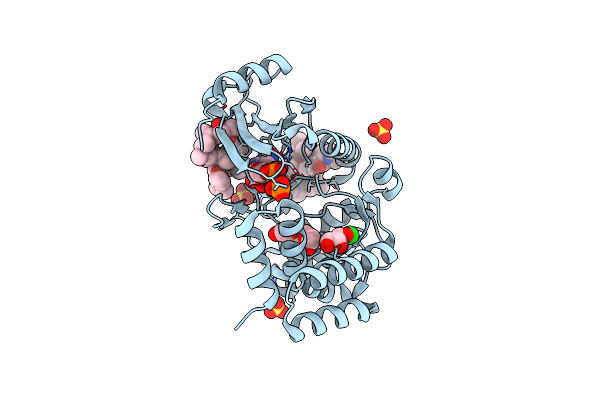
Crystal Structure Of Macrolide 2'-Phosphotransferase Mphh From Brachybacterium Faecium In Complex With Azithromycin
Organism: Brachybacterium faecium (strain atcc 43885 / dsm 4810 / ncib 9860)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-08-23 Classification: TRANSFERASE/ANTIBIOTIC Ligands: ZIT, SO4, CL |
 |
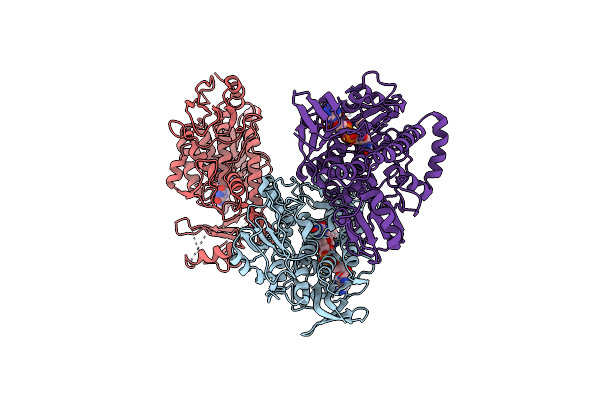
Crystal Structure Of Macrolide 2'-Phosphotransferase Mphh From Brachybacterium Faecium In Complex With Gdp
Organism: Brachybacterium faecium
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2017-08-16 Classification: TRANSFERASE/ANTIBIOTIC Ligands: MG, GDP, SO4, ZIT, PO4, CL, PGE, GOL |
 |
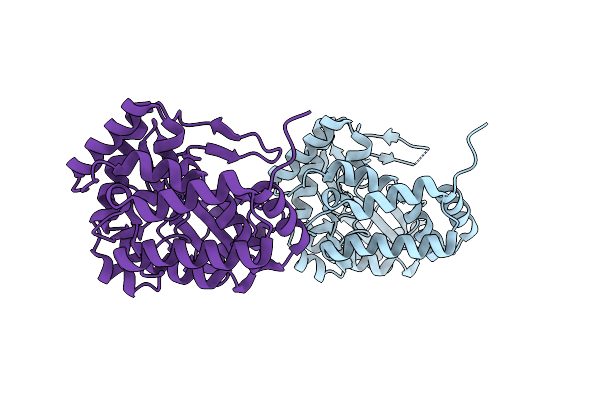
Crystal Structure Of Rifampin Monooxygenase From Streptomyces Venezuelae, Complex With Fad
Organism: Streptomyces venezuelae (strain atcc 10712 / cbs 650.69 / dsm 40230 / jcm 4526 / nbrc 13096 / pd 04745)
Method: X-RAY DIFFRACTION Resolution:3.39 Å Release Date: 2017-08-16 Classification: OXIDOREDUCTASE Ligands: FAD, CL, GOL |
 |
Crystal Structure Of Macrolide 2'-Phosphotransferase Mphh From Brachybacterium Faecium, Apoenzyme
Organism: Brachybacterium faecium (strain atcc 43885 / dsm 4810 / ncib 9860)
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2017-07-26 Classification: TRANSFERASE Ligands: CL |

