Search Count: 16
 |
Crystal Structure Of Arabidopsis Thaliana Udp-Glucose 4-Epimerase 2 (Atuge2) Complexed With Udp, G233A Mutant
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2024-05-15 Classification: ISOMERASE Ligands: NAD, UDP |
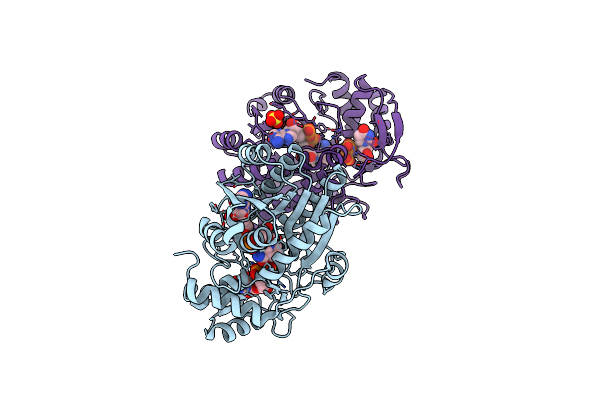 |
Crystal Structure Of Arabidopsis Thaliana Udp-Glucose 4-Epimerase 2 (Atuge2) Complexed With Udp, I160L/G233A Mutant
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-05-15 Classification: ISOMERASE Ligands: UDP, NAD, SO4 |
 |
Crystal Structure Of Arabidopsis Thaliana Udp-Glucose 4-Epimerase 2 (Atuge2) Complexed With Udp, Wild-Type
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-05-08 Classification: ISOMERASE Ligands: UDP, NAD |
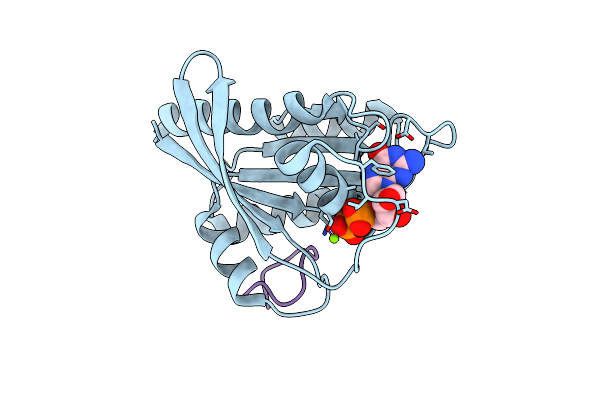 |
Human K-Ras G12D (Gdp-Bound) In Complex With Cyclic Peptide Inhibitor Ap8784
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2023-07-26 Classification: SIGNALING PROTEIN/INHIBITOR Ligands: GDP, MG, IOD |
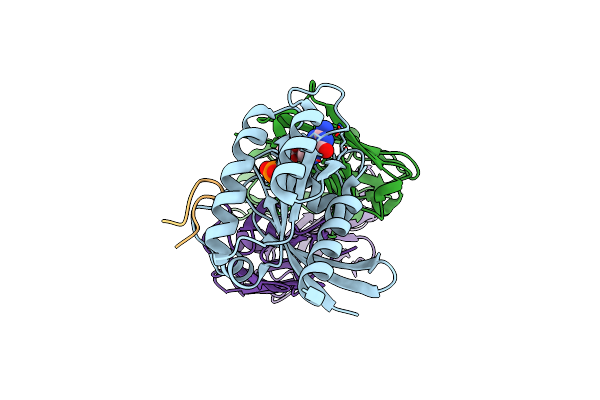 |
Human K-Ras G12D (Gdp-Bound) In Complex With Cyclic Peptide Inhibitor Luna18 And Ka30L Fab
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2023-07-26 Classification: SIGNALING PROTEIN Ligands: GDP, MG |
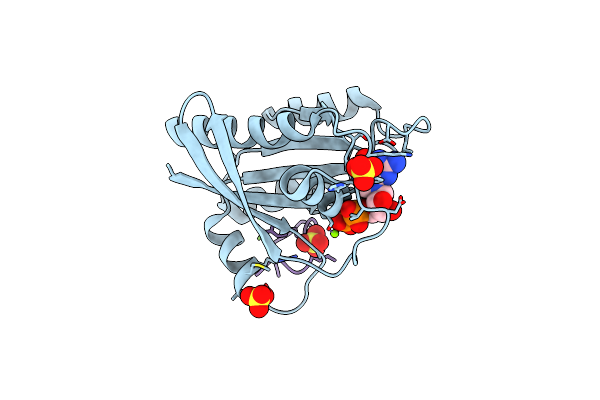 |
Human K-Ras G12D (Gdp-Bound) In Complex With Cyclic Peptide Inhibitor Ap10343
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2023-07-26 Classification: SIGNALING PROTEIN/INHIBITOR Ligands: GDP, MG, SO4 |
 |
Crystal Structure Of A Novel 4-O-Alpha-L-Rhamnosyl-Beta-D-Glucuronidase From Fusarium Oxysporum 12S, Ligand-Free Form
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2021-03-17 Classification: HYDROLASE Ligands: NAG |
 |
Crystal Structure Of A Novel 4-O-Alpha-L-Rhamnosyl-Beta-D-Glucuronidase From Fusarium Oxysporum 12S - Rha-Glca Complex
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2021-03-17 Classification: HYDROLASE Ligands: NAG, MAN |
 |
Crystal Structure Of Exo-Beta-1,3-Galactanase From Phanerochaete Chrysosporium Pc1,3Gal43A Apo Form
Organism: Phanerochaete chrysosporium
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-11-04 Classification: HYDROLASE Ligands: NAG, CA, CIT |
 |
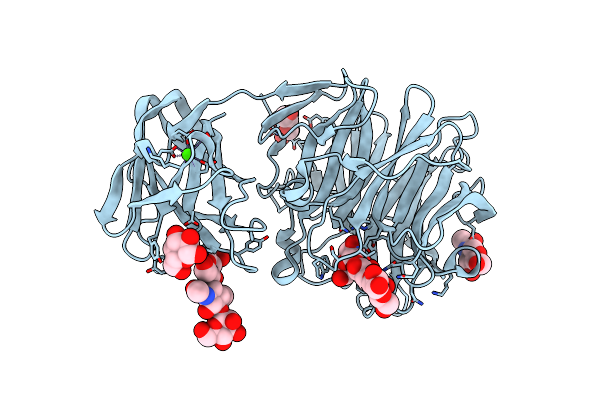
Crystal Structure Of Exo-Beta-1,3-Galactanase From Phanerochaete Chrysosporium Pc1,3Gal43A With Galactose
Organism: Phanerochaete chrysosporium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-11-04 Classification: HYDROLASE Ligands: NAG, CA, GLA, GAL, ACY, ACT, GOL |
 |
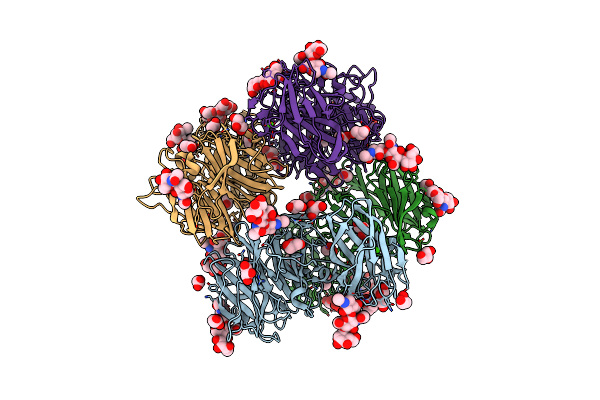
Crystal Structure Of Exo-Beta-1,3-Galactanase From Phanerochaete Chrysosporium Pc1,3Gal43A E208Q With Beta-1,3-Galactotriose
Organism: Phanerochaete chrysosporium
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-11-04 Classification: HYDROLASE Ligands: NAG, CA, GAL |
 |
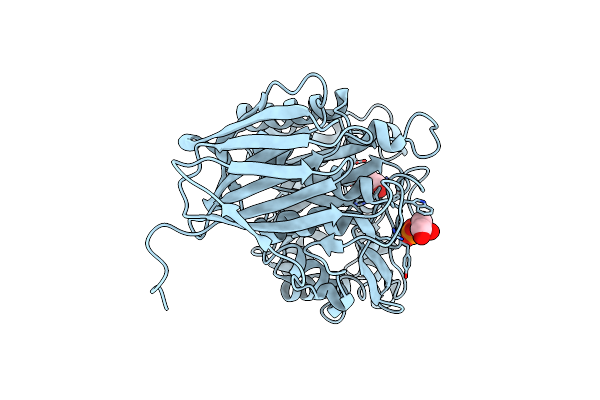
Crystal Structure Of Exo-Beta-1,3-Galactanase From Phanerochaete Chrysosporium Pc1,3Gal43A E208A With Beta-1,3-Galactotriose
Organism: Phanerochaete chrysosporium
Method: X-RAY DIFFRACTION Release Date: 2020-11-04 Classification: HYDROLASE Ligands: CA, ACT, GOL, NAG |
 |
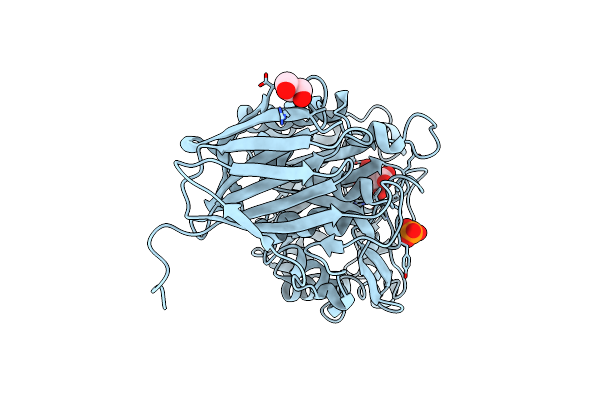
Organism: Acidobacterium capsulatum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-02-22 Classification: HYDROLASE Ligands: PO4, GOL |
 |
Crystal Structure Of Beta-Glucuronidase From Acidobacterium Capsulatum In Complex With D-Glucuronic Acid
Organism: Acidobacterium capsulatum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-02-22 Classification: HYDROLASE Ligands: BDP, PO4, GOL |
 |
Crystal Structure Of Beta-Glucuronidase From Acidobacterium Capsulatum Covalent-Bonded With 2-Deoxy-2-Fluoro-D-Glucuronic Acid
Organism: Acidobacterium capsulatum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-02-22 Classification: HYDROLASE Ligands: GUZ, GUF, DNF, PO4 |
 |
Crystal Structure Of Udp-Galactopyranose Mutase In Complex With Phosphonate Analog Of Udp-Galactopyranose
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2010-10-20 Classification: ISOMERASE Ligands: FAD, UDP, GOL, URM, XYL |

