Search Count: 40
 |
Organism: Bacillus thuringiensis ybt-1518
Method: X-RAY DIFFRACTION Resolution:4.50 Å Release Date: 2022-12-07 Classification: TOXIN |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-05-25 Classification: HYDROLASE |
 |
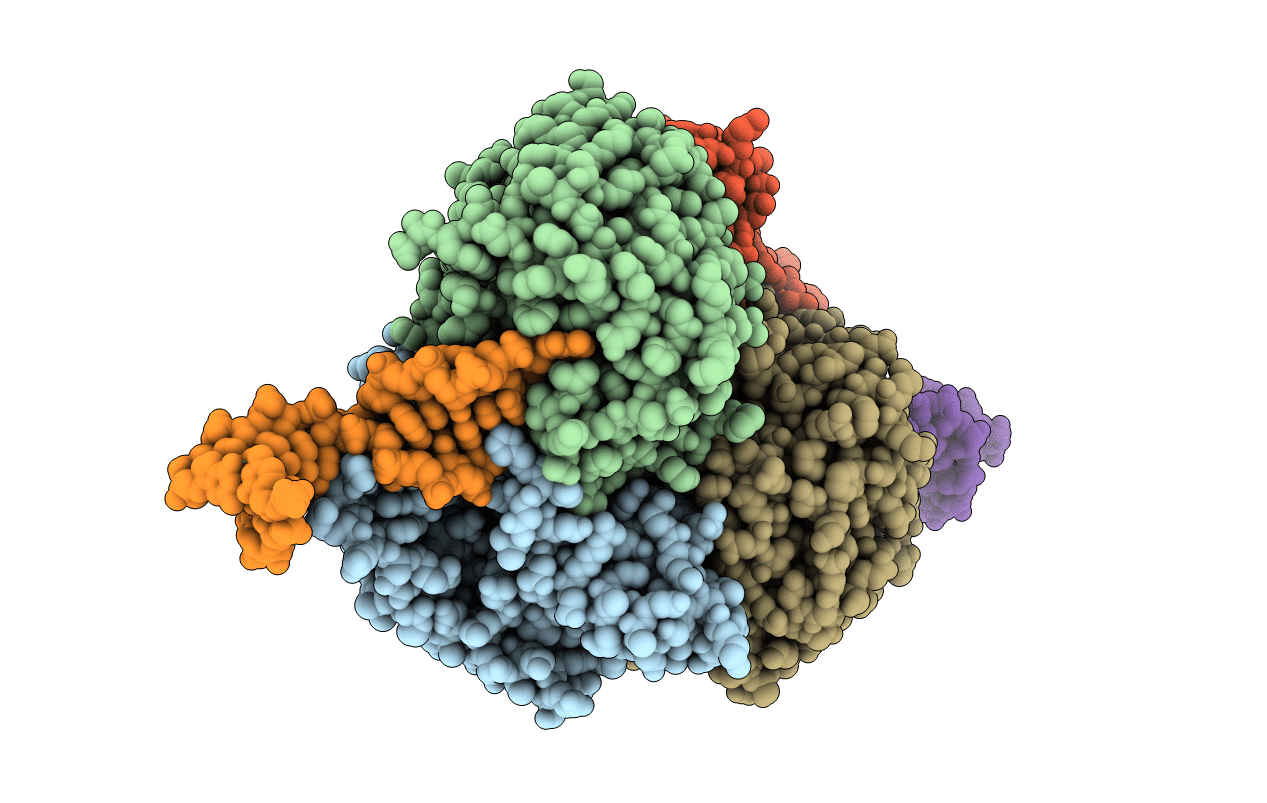
Porcine Cd38 Complexed With Complexed With A Covalent Intermediate, Ribo-F-Ribose-5'-Phosphate
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-05-25 Classification: HYDROLASE Ligands: AVW |
 |
Structure Of Plasmodium Falciparum Lactate Dehydrogenase In Complex With A Dna Aptamer
Organism: Plasmodium falciparum, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-09-25 Classification: OXIDOREDUCTASE |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2013-05-08 Classification: MOTOR PROTEIN |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-05-08 Classification: MOTOR PROTEIN |
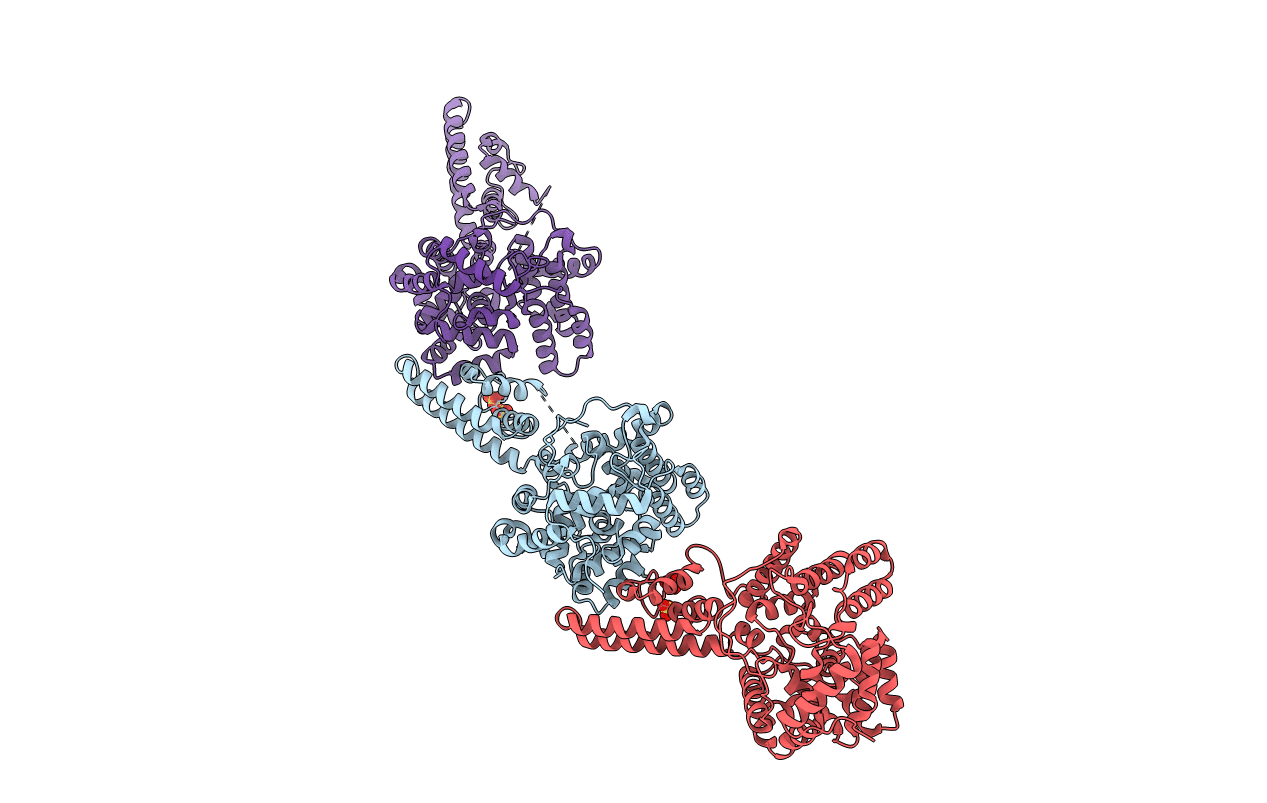 |
X-Ray Crystallographic Structure Of Crimean-Congo Haemorrhagic Fever Virus Nucleoprotein
Organism: Crimean-congo hemorrhagic fever virus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2012-09-26 Classification: VIRAL PROTEIN Ligands: SO4 |
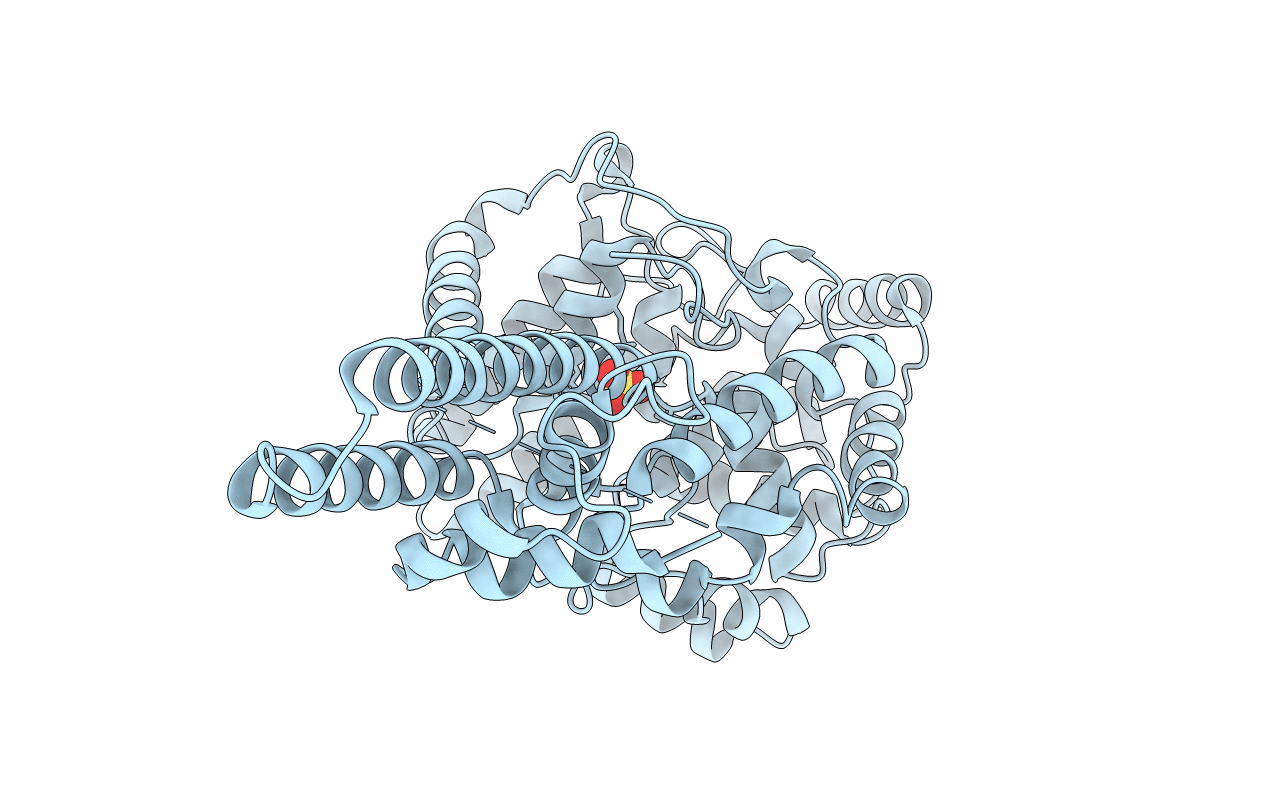 |
X-Ray Crystallographic Structure Of Crimean-Congo Haemorrhagic Fever Virus Nucleoprotein
Organism: Crimean-congo hemorrhagic fever virus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2012-09-26 Classification: VIRAL PROTEIN Ligands: SO4 |
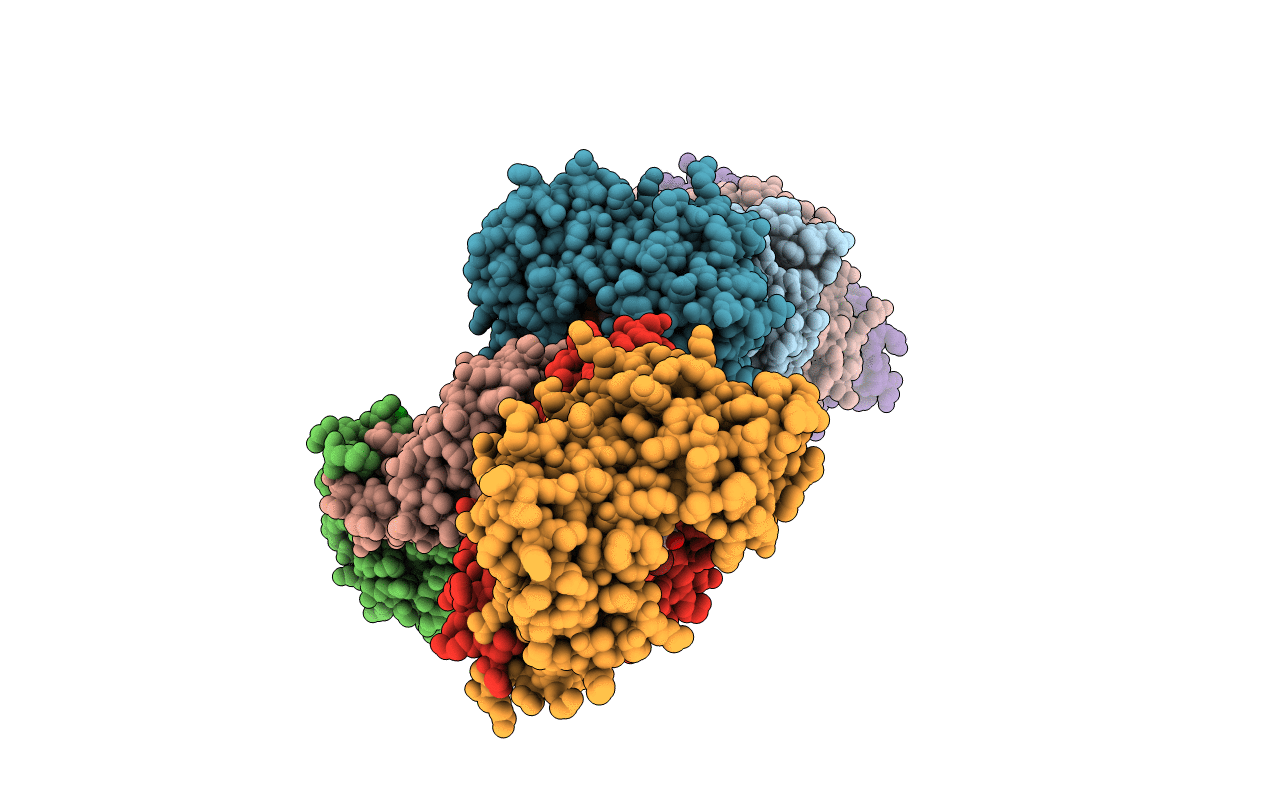 |
Organism: Aplysia californica
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-08-22 Classification: HYDROLASE Ligands: CL, AV1 |
 |
Crystal Structure Of Adp-Ribosyl Cyclase Complexed With 8-Bromo-Adp- Ribose And Cyclic 8-Bromo-Cyclic-Adp-Ribose
Organism: Aplysia californica
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2012-08-22 Classification: HYDROLASE Ligands: AV1, CV1 |
 |
Crystal Structure Of The N-Terminal Domain Of The Human Dead-Box Rna Helicase Ddx5 (P68)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2012-08-08 Classification: HYDROLASE |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-07-18 Classification: TRANSCRIPTION REGULATOR Ligands: MG, EPE |
 |
Crystal Structure Of Adp Ribosyl Cyclase Complexed With Substrate Nad And Product Cadpr
Organism: Aplysia californica
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-11-30 Classification: HYDROLASE Ligands: NAD, CXR |
 |
Crystal Structure Of Aplysia Cyclase Complexed With Substrate Ngd And Product Cgdpr
Organism: Aplysia californica
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-11-30 Classification: HYDROLASE Ligands: CGR, NGD |
 |
Organism: Aplysia californica
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-11-30 Classification: HYDROLASE Ligands: G2Q, NGD |
 |
Crystal Structure Of Adp Ribosyl Cyclase Complexed With Ara-2'F-Adp- Ribose At 2.1 Angstrom
Organism: Aplysia californica
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2011-11-30 Classification: HYDROLASE Ligands: AVU, GOL |
 |
Crystal Structure Of Adp-Ribosyl Cyclase Complexed With Ara-2'F-Adp- Ribose At 2.3 Angstrom
Organism: Aplysia californica
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-11-30 Classification: HYDROLASE Ligands: AVU |
 |
Crystal Structure Of Adp-Ribosyl Cyclase Complexed With Ara-2'F-Adp- Ribose At 2.3 Angstrom
Organism: Aplysia californica
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-11-30 Classification: HYDROLASE Ligands: AVU |
 |
Induced-Fit And Allosteric Effects Upon Polyene Binding Revealed By Crystal Structures Of The Dynemicin Thioesterase
Organism: Micromonospora chersina
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-10-13 Classification: BIOSYNTHETIC PROTEIN Ligands: SSV |
 |
Induced-Fit And Allosteric Effects Upon Polyene Binding Revealed By Crystal Structures Of The Dynemicin Thioesterase
Organism: Micromonospora chersina
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2010-10-13 Classification: HYDROLASE |

