Search Count: 14
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2025-04-23 Classification: DE NOVO PROTEIN Ligands: NH4, SO4, ACT, TRS |
 |
Organism: Enterococcus hirae atcc 9790
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2023-07-12 Classification: MOTOR PROTEIN |
 |
Hexameric Ring Complex Of Engineered V1-Atpase Bound To 5 Adps: A3(De)3_(Adp-Pi)1Cat(Adp)2Cat,2Non-Cat
Organism: Enterococcus hirae atcc 9790
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2023-07-12 Classification: MOTOR PROTEIN Ligands: ADP, MG, PO4 |
 |
Hexameric Ring Complex Of Engineered V1-Atpase Bound To 4 Adps: A3(De)3_(Adp)3Cat,1Non-Cat, Hexameric Ring Complex Of Engineered V1-Atpase Bound To 5 Adps: A3(De)3_(Adp)3Cat,2Non-Cat
Organism: Enterococcus hirae atcc 9790
Method: X-RAY DIFFRACTION Resolution:4.20 Å Release Date: 2023-07-12 Classification: MOTOR PROTEIN Ligands: ADP, MG |
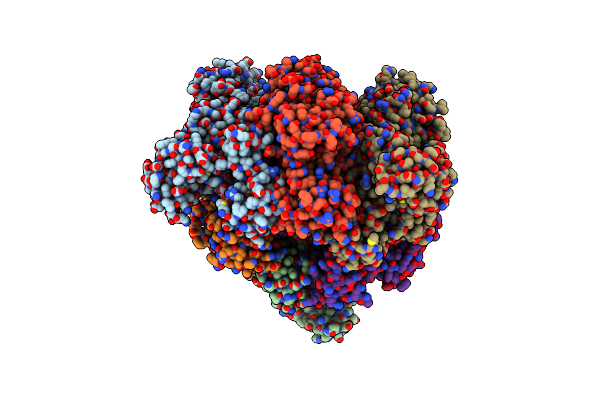 |
Hexameric Ring Complex Of Engineered V1-Atpase Bound To Amp-Pnp: A3(De)3_(Anp)1Cat
Organism: Enterococcus hirae atcc 9790
Method: X-RAY DIFFRACTION Resolution:3.44 Å Release Date: 2021-08-11 Classification: MOTOR PROTEIN Ligands: MG, ANP |
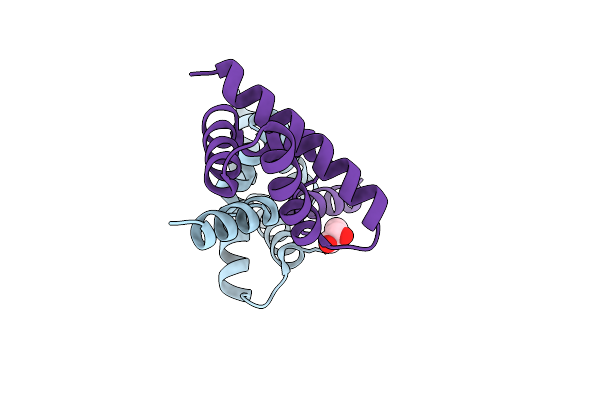 |
Crystal Structure Of Domain-Swapped Dimer Of H5_Fold-0 Elsa; De Novo Designed Protein With An Asymmetric All-Alpha Topology
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2021-07-28 Classification: DE NOVO PROTEIN Ligands: GOL |
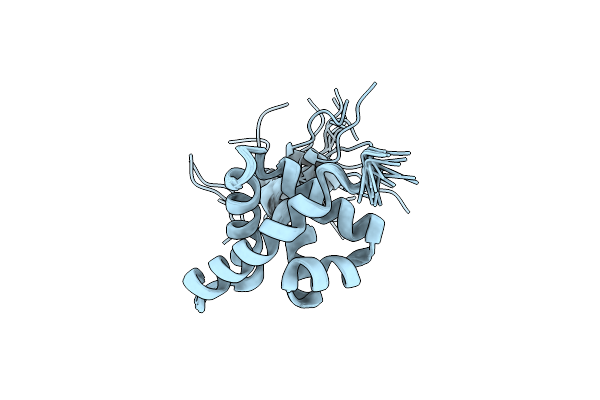 |
Solution Nmr Structure Of Fold-0 Chantal; De Novo Designed Protein With An Asymmetric All-Alpha Topology
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2021-04-07 Classification: DE NOVO PROTEIN |
 |
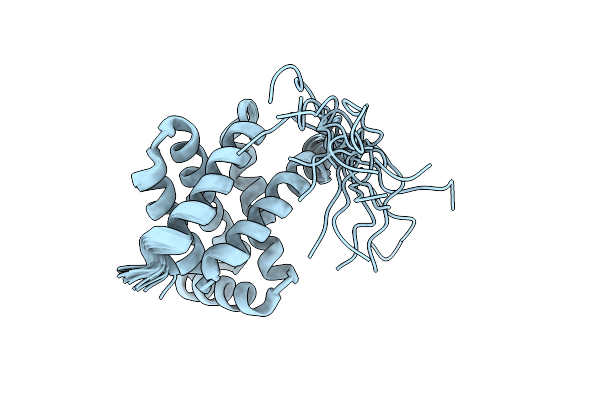
Solution Nmr Structure Of Fold-C Rei; De Novo Designed Protein With An Asymmetric All-Alpha Topology
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2021-04-07 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of Fold-Z Gogy; De Novo Designed Protein With An Asymmetric All-Alpha Topology
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2021-04-07 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of Fold-K Mussoc; De Novo Designed Protein With An Asymmetric All-Alpha Topology
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2021-04-07 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of Fold-U Nomur; De Novo Designed Protein With An Asymmetric All-Alpha Topology
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2021-04-07 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of De Novo Rossmann2X2 Fold With Most Of The Core Mutated To Valine, R2X2_Val88
Organism: Unidentified
Method: SOLUTION NMR Release Date: 2020-12-02 Classification: DE NOVO PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-10-30 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: ACT, PDY |
 |
Crystal Structure Of A Pyrazolopyrimidine Inhibitor Complex Bound To Mapkap Kinase-2 (Mk2)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2010-05-12 Classification: TRANSFERASE Ligands: PDY, SO4 |

