Search Count: 27
 |
Organism: Bacillus cereus vd154
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: METAL BINDING PROTEIN Ligands: ZN, MG |
 |
Organism: Lama glama, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2022-12-28 Classification: IMMUNE SYSTEM/INHIBITOR Ligands: GOL, CL, SO4 |
 |
Organism: Lama glama
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2020-05-13 Classification: IMMUNE SYSTEM |
 |
Organism: Lama glama
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-07-24 Classification: IMMUNE SYSTEM Ligands: SO4, CL |
 |
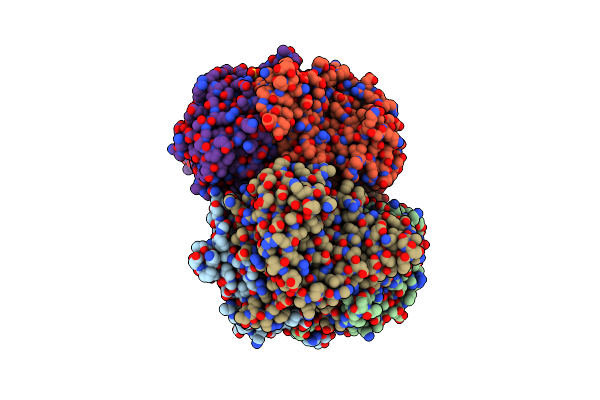
Crystal Structure Of Two-Domain Laccase Mutant I170F From Streptomyces Griseoflavus
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-07-17 Classification: OXIDOREDUCTASE Ligands: CU, EDO, SO4, GOL, OXY |
 |
Crystal Structure Of Two-Domain Laccase Mutant I170A From Streptomyces Griseoflavus
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2019-07-17 Classification: OXIDOREDUCTASE Ligands: CU, GOL, SO4 |
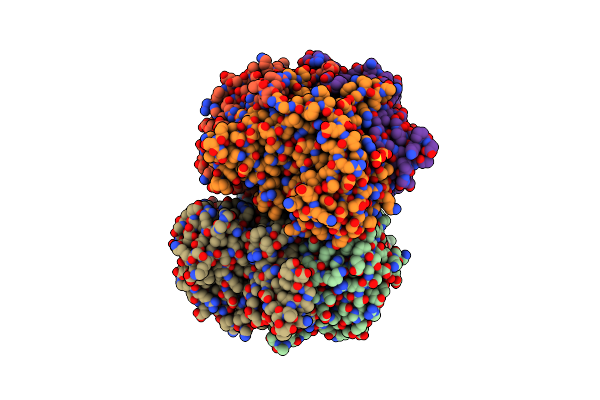 |
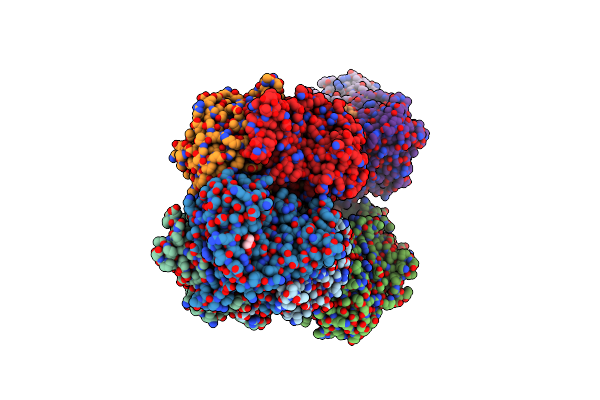
Crystal Structure Of Two-Domain Laccase From Streptomyces Griseoflavus Produced At 0.25 Mm Copper Sulfate In Growth Medium
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-07-17 Classification: OXIDOREDUCTASE Ligands: CU, PEO, EDO, GOL, SO4 |
 |
Crystal Structure Of Two-Domain Laccase Mutant H165F From Streptomyces Griseoflavus With High Copper Ions Occupancy
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-01-30 Classification: OXIDOREDUCTASE Ligands: CU, PER, GOL, OXY |
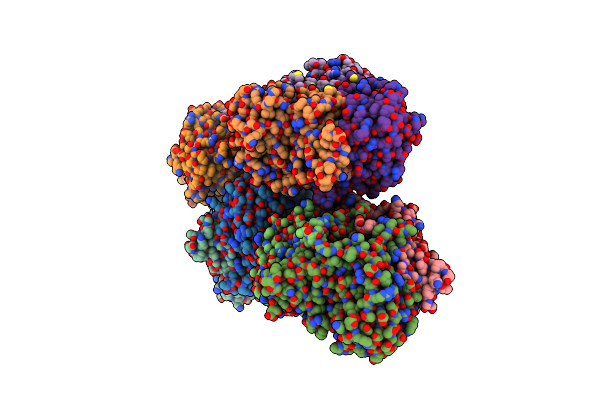 |
Crystal Structure Of Two-Domain Laccase Mutant H165A From Streptomyces Griseoflavus With High Copper Ions Occupancy
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2019-01-30 Classification: OXIDOREDUCTASE Ligands: CU, GOL, OXY |
 |
Crystal Structure Of Mutant Ribosomal Protein Tthl1 Lacking 8 N-Terminal Residues In Complex With 80Nt 23S Rna From Thermus Thermophilus
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-05-16 Classification: RNA Ligands: MLI, NA |
 |
Crystal Structure Of Mutant M54L/M64L/M96L Of Two-Domain Laccase From Streptomyces Griseoflavus With 1 Mm Copper Sulfate On Growth Medium
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-03-28 Classification: OXIDOREDUCTASE Ligands: CU, O, GOL, OXY |
 |
Crystal Structure Of Mutant M54L/M64L/M96L Of Two-Domain Laccase From Streptomyces Griseoflavus Dialyzed Against Solution Containing 0.25 Mm Copper Sulfate
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-03-28 Classification: OXIDOREDUCTASE Ligands: CU, GOL, SO4 |
 |
Crystal Structure Of Mutant M54L/M64L/M96L Of Two-Domain Laccase From Streptomyces Griseoflavus With 0.25 Mm Copper Sulfate On Growth Medium
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-03-28 Classification: OXIDOREDUCTASE Ligands: CU, TRS, AZI, GOL, FMT, SO4 |
 |
Crystal Structure Of Two-Domain Laccase Mutant H165F From Streptomyces Griseoflavus
Organism: Streptomyces griseoflavus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-12-20 Classification: OXIDOREDUCTASE Ligands: CU, OXY, O, EDO, GOL, SO4 |
 |
Mutant Ribosomal Protein L1 From Thermus Thermophilus With Threonine 217 Replaced By Valine
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2015-03-18 Classification: RNA BINDING PROTEIN Ligands: SO4, MPD, GLY |
 |
Crystal Structure Of Mutant Ribosomal Protein G219V Tthl1 In Complex With 80Nt 23S Rna From Thermus Thermophilus
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-02-11 Classification: RIBOSOMAL PROTEIN/RNA Ligands: SO4, IPA, BME, URE |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-02-11 Classification: RIBOSOMAL PROTEIN Ligands: CL, ACT |
 |
Crystal Structure Of Mutant Ribosomal Protein M218L Tthl1 In Complex With 80Nt 23S Rna From Thermus Thermophilus
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-02-11 Classification: RIBOSOMAL PROTEIN/RNA Ligands: SO4, ACT, MES |
 |
Crystal Structure Of Mutant Ribosomal Protein L1 From Methanococcus Jannaschii With Deletion Of 8 Residues From C-Terminus
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-07-02 Classification: RIBOSOMAL PROTEIN Ligands: TLA, IPA, CL |
 |
Crystal Structure Of Mutant Ribosomal Protein T217V Tthl1 In Complex With 80Nt 23S Rna From Thermus Thermophilus
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-10-31 Classification: RNA/RNA BINDING PROTEIN Ligands: MG, MLI |

