Search Count: 63
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2018-03-07 Classification: HYDROLASE Ligands: ZN, BCN |
 |
Crystal Structure Of Metallo-Beta-Lactamase Vim-1 In Complex With Ml302F Inhibitor
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2018-03-07 Classification: HYDROLASE Ligands: ZN, S3C |
 |
Crystal Structure Of Vim-1 Metallo-Beta-Lactamase In Complex With Hydrolysed Meropenem
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-03-07 Classification: HYDROLASE Ligands: ZN, LMP |
 |
Crystal Structure Of Metallo-Beta-Lactamase Spm-1 Complexed With Cyclobutanone Inhibitor
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2018-01-17 Classification: HYDROLASE Ligands: ZN, GOL, DMS, CL, 8TW, SO4, TRS |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-01-17 Classification: HYDROLASE Ligands: ZN, SO4, GOL, CL |
 |
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2016-06-01 Classification: HYDROLASE Ligands: ZN, EDO, ACT |
 |
Crystal Structure Of The Metallo-Beta-Lactamase Imp-1 In Complex With The Bisthiazolidine Inhibitor D-Cs319
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-06-01 Classification: HYDROLASE Ligands: ZN, 3R9, EDO, DMS, NA, CL |
 |
Crystal Structure Of The Metallo-Beta-Lactamase L1 In Complex With The Bisthiazolidine Inhibitor D-Cs319
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2016-06-01 Classification: HYDROLASE Ligands: ZN, 3R9, SO4 |
 |
Crystal Structure Of The Metallo-Beta-Lactamase L1 In Complex With The Bisthiazolidine Inhibitor D-Vc26
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-06-01 Classification: HYDROLASE Ligands: VC2, ZN, SO4 |
 |
Crystal Structure Of The Metallo-Beta-Lactamase L1 In Complex With The Bisthiazolidine Inhibitor L-Cs319
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2016-06-01 Classification: HYDROLASE Ligands: 3C7, SO4, ZN |
 |
Crystal Structure Of The Metallo-Beta-Lactamase Sfh-I In Complex With The Bisthiazolidine Inhibitor L-Cs319
Organism: Serratia fonticola
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2016-06-01 Classification: HYDROLASE Ligands: ZN, 3C7 |
 |
Crystal Structure Of The Metallo-Beta-Lactamase Imp-1 In Complex With The Bisthiazolidine Inhibitor L-Vc26
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-06-01 Classification: HYDROLASE Ligands: ZN, 9BZ, EDO, DMS |
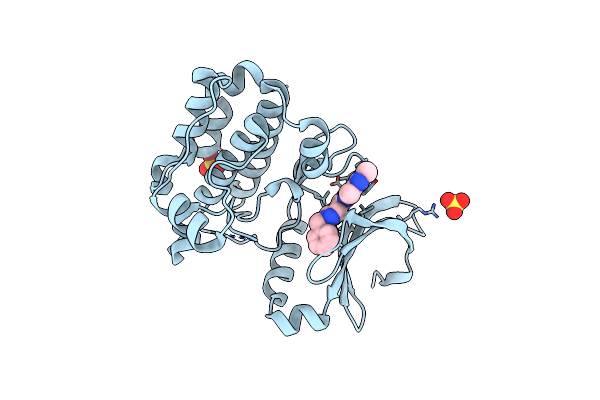 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2015-09-02 Classification: TRANSFERASE Ligands: 5GX, SO4 |
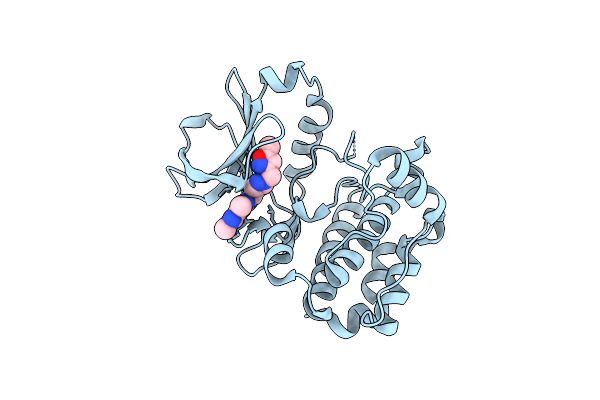 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2015-09-02 Classification: TRANSFERASE Ligands: 7HD |
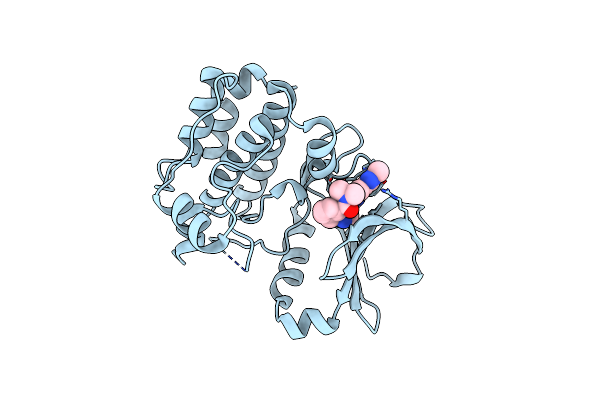 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2015-09-02 Classification: TRANSFERASE Ligands: NL4 |
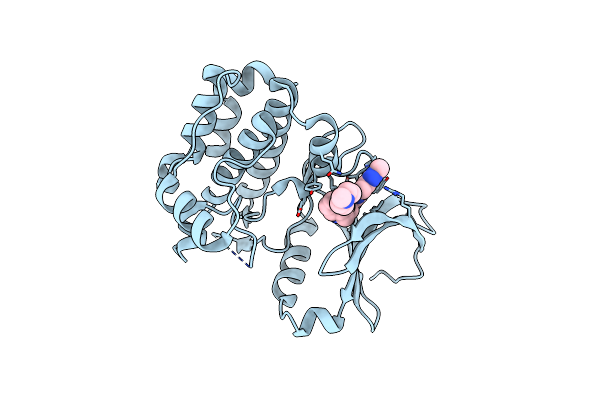 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2015-09-02 Classification: TRANSFERASE Ligands: 6F2 |
 |
Organism: Pseudomonas stutzeri
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2015-05-27 Classification: HYDROLASE Ligands: ZN, CL |
 |
Bacillus Cereus Zn-Dependent Metallo-Beta-Lactamase At Ph 7 Complexed With Compound L-Cs319
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-11-26 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, K, 3R9 |
 |
Organism: Pseudomonas stutzeri
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-09-17 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of The Metallo-Beta-Lactamase Ndm-1 In Complex With A Bisthiazolidine Inhibitor
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-08-20 Classification: HYDROLASE Ligands: ZN, 3C7, GOL |

