Search Count: 23
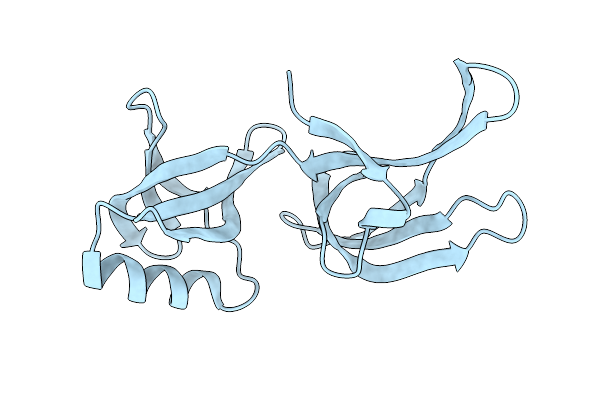 |
Structure Of Hex-1 From N. Crassa Crystallized In Cellulo (Cytosol), Diffracted At 100K And Resolved Using Crystfel
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2024-02-21 Classification: STRUCTURAL PROTEIN |
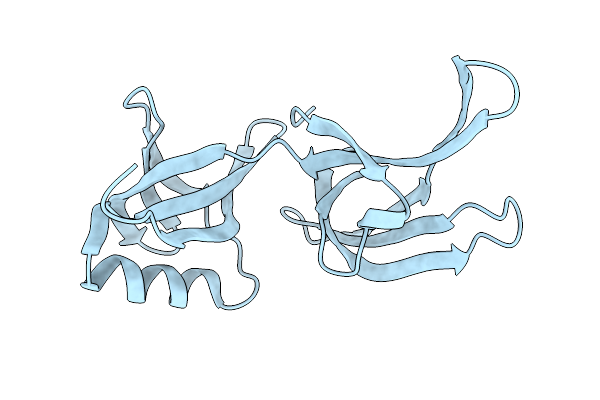 |
Structure Of Hex-1 From N. Crassa Crystallized In Cellulo, Diffracted At 100K And Resolved Using Crystfel
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2024-02-21 Classification: STRUCTURAL PROTEIN |
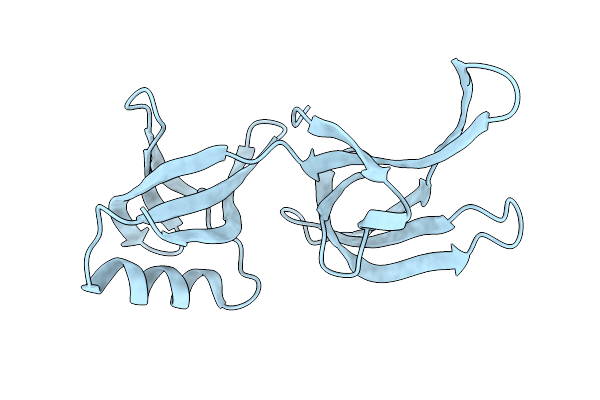 |
Structure Of Hex-1 (Cyto V2) From N. Crassa Grown In Living Insect Cells, Diffracted At 100K And Resolved Using Crystfel
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-02-21 Classification: STRUCTURAL PROTEIN |
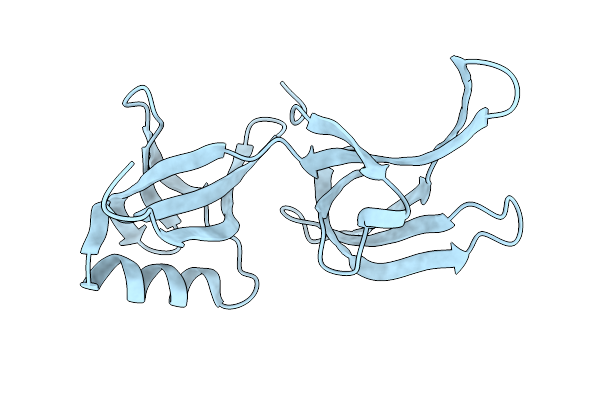 |
Structure Of Hex-1 From N. Crassa Crystallized In Cellulo, Diffracted At 100K And Resolved Using Xds
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-02-21 Classification: STRUCTURAL PROTEIN |
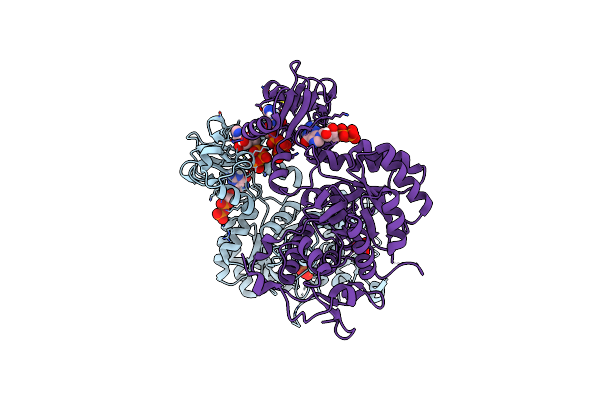 |
Trypanosoma Brucei Imp Dehydrogenase (Ori) Crystallized In High Five Cells Reveals Native Ligands Atp, Gdp And Phosphate. Diffraction Data Collection At 100 K In Cellulo; Xds Processing
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-02-21 Classification: TRANSFERASE Ligands: ATP, GDP, PO4 |
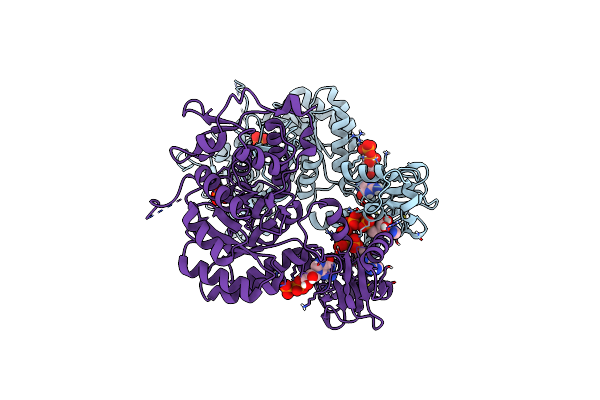 |
Trypanosoma Brucei Imp Dehydrogenase (Cyto) Crystallized In High Five Cells Revealing Native Ligands Atp, Gdp And Phosphate. Diffraction Data Collection At 100 K In Cellulo
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-01-17 Classification: TRANSFERASE Ligands: GDP, ATP, PO4 |
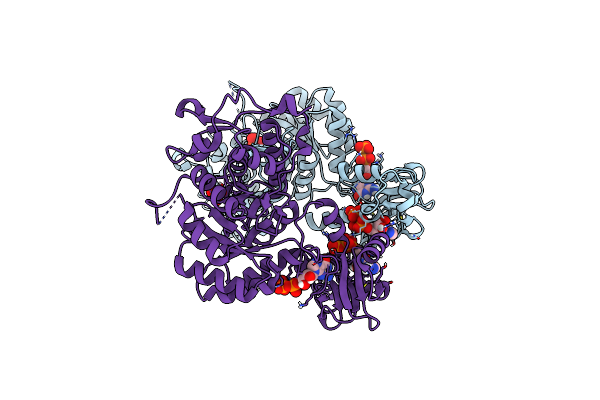 |
Trypanosoma Brucei Imp Dehydrogenase (Ori) Crystallized In High Five Cells Reveals Native Ligands Atp, Gdp And Phosphate. Diffraction Data Collection At 100 K In Cellulo; Crystfel Processing
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-01-17 Classification: TRANSFERASE Ligands: GDP, ATP, PO4 |
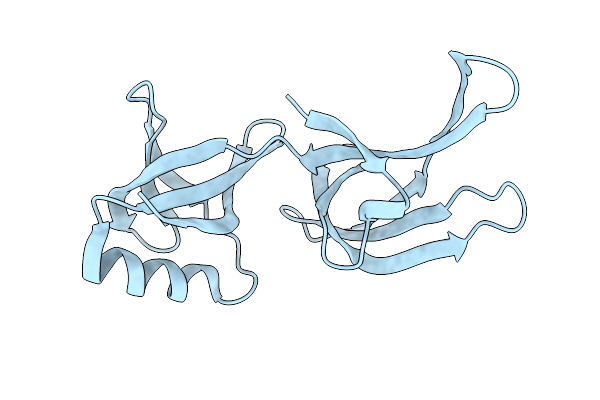 |
Hex-1 (In Cellulo, In Situ) Crystallized And Diffracted In High Five Cells. Growth And Sx Data Collection At 296 K On Crystaldirect Plates
Organism: Neurospora crassa or74a
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2024-01-17 Classification: STRUCTURAL PROTEIN |
 |
Crystal Structure Of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed With Inhibitor Mz317
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2023-12-13 Classification: HYDROLASE/INHIBITOR Ligands: YIZ, ZN, K, EDO |
 |
Crystal Structure Of Danio Rerio Histone Deacetylase 6 Catalytic Domain 2 Complexed With Inhibitor Mz327
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2023-12-13 Classification: HYDROLASE/INHIBITOR Ligands: YJ5, EDO, ZN, K, EOH |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-11-15 Classification: HYDROLASE Ligands: KZU, EDO, BTB, BU3, ZN, CL |
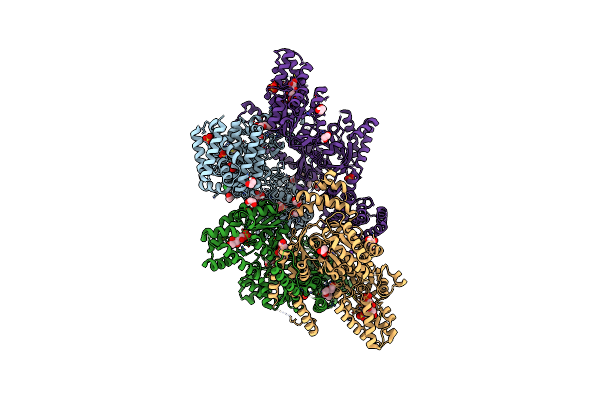 |
Mannitol-1-Phosphate Bound To The Phosphatase Domain Of The Bifunctional Mannitol-1-Phosphate Dehydrogenase/Phosphatase Mtld-D16A From Acinetobacter Baumannii
Organism: Acinetobacter baumannii atcc 19606 = cip 70.34 = jcm 6841
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-04-27 Classification: OXIDOREDUCTASE Ligands: GOL, EDO, PGE, PG4, BME, SO4, MG, CL, 44H, EPE, NA, ACT |
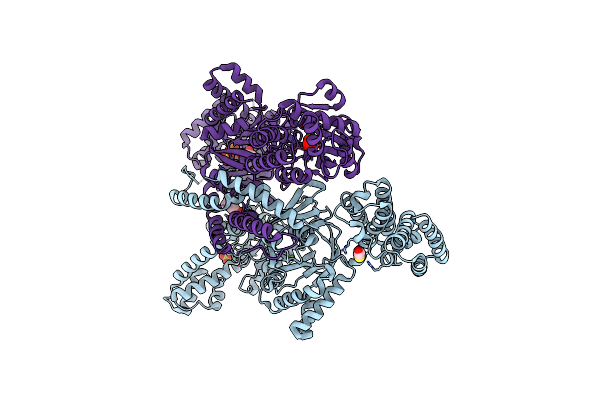 |
Mannitol-1-Phosphate Bound To The Phosphatase Domain Of The Bifunctional Mannitol-1-Phosphate Dehydrogenase/Phosphatase Mtld-N374A From Acinetobacter Baumannii
Organism: Acinetobacter baumannii atcc 19606 = cip 70.34 = jcm 6841
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-04-27 Classification: OXIDOREDUCTASE Ligands: BME, MG, SO4, CL, EPE, 44H |
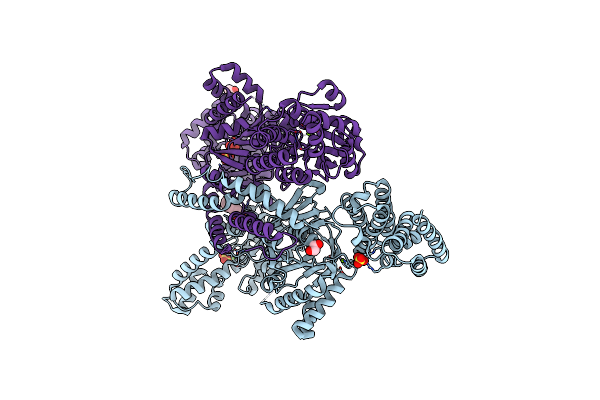 |
Crystal Structure Of The Bifunctional Mannitol-1-Phosphate Dehydrogenase/Phosphatase Mtld From Acinetobacter Baumannii
Organism: Acinetobacter baumannii (strain atcc 19606 / dsm 30007 / cip 70.34 / jcm 6841 / nbrc 109757 / ncimb 12457 / nctc 12156 / 81)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-04-20 Classification: OXIDOREDUCTASE Ligands: MG, SO4, EDO, CL, BR, EPE |
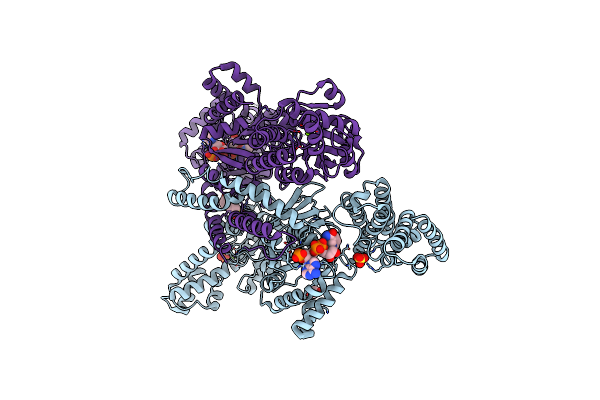 |
Nadph Bound To The Dehydrogenase Domain Of The Bifunctional Mannitol-1-Phosphate Dehydrogenase/Phosphatase Mtld From Acinetobacter Baumannii
Organism: Acinetobacter baumannii (strain atcc 19606 / dsm 30007 / cip 70.34 / jcm 6841 / nbrc 109757 / ncimb 12457 / nctc 12156 / 81)
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2022-04-20 Classification: OXIDOREDUCTASE Ligands: EDO, MG, SO4, NDP, CL, EPE |
 |
Nadh Bound To The Dehydrogenase Domain Of The Bifunctional Mannitol-1-Phosphate Dehydrogenase/Phosphatase Mtld From Acinetobacter Baumannii
Organism: Acinetobacter baumannii (strain atcc 19606 / dsm 30007 / cip 70.34 / jcm 6841 / nbrc 109757 / ncimb 12457 / nctc 12156 / 81)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-04-20 Classification: OXIDOREDUCTASE Ligands: V8H, MG, SO4, EDO, GOL, CL, EPE, NAI |
 |
Nadph And Fructose-6-Phosphate Bound To The Dehydrogenase Domain Of The Bifunctional Mannitol-1-Phosphate Dehydrogenase/Phosphatase Mtld From Acinetobacter Baumannii
Organism: Acinetobacter baumannii (strain atcc 19606 / dsm 30007 / cip 70.34 / jcm 6841 / nbrc 109757 / ncimb 12457 / nctc 12156 / 81)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-04-20 Classification: OXIDOREDUCTASE Ligands: SO4, NDP, EPE, EDO, CL, MG, K, F6R |
 |
Nadph Bound To The Dehydrogenase Domain Of The Bifunctional Mannitol-1-Phosphate Dehydrogenase/Phosphatase Mtld-N374A From Acinetobacter Baumannii
Organism: Acinetobacter baumannii (strain atcc 19606 / dsm 30007 / cip 70.34 / jcm 6841 / nbrc 109757 / ncimb 12457 / nctc 12156 / 81)
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2022-04-20 Classification: OXIDOREDUCTASE Ligands: EDO, MG, NDP, SO4, CL, K, EPE, ACT |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2002-04-17 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2002-03-06 Classification: DNA BINDING PROTEIN |

