Search Count: 22
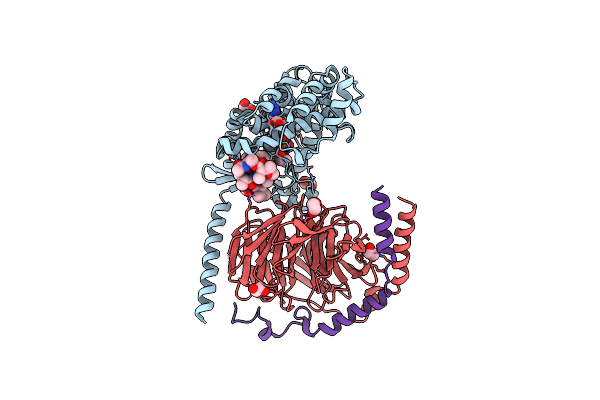 |
Crystal Structure Of The G11 Protein Heterotrimer Bound To Ym-254890 Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-03-19 Classification: SIGNALING PROTEIN Ligands: GDP, EDO, DAM, HF2, THC, OTH, HL2, MAA, ALA, ACE |
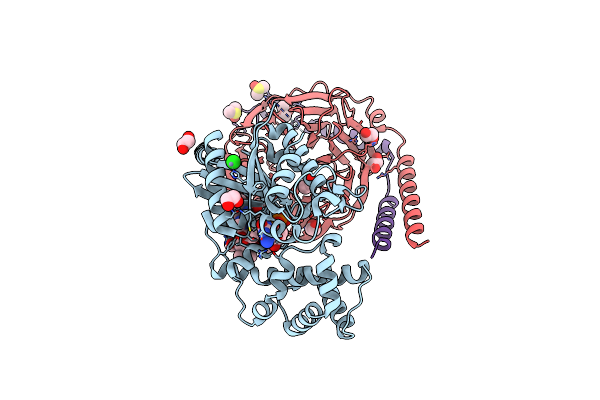 |
Crystal Structure Of The G11 Protein Heterotrimer Bound To Fr900359 Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2025-03-19 Classification: SIGNALING PROTEIN Ligands: EDO, GDP, ZN, CL, DAM, HF2, UDL, OTH, HL2, MAA, ALA, PPI, DMS |
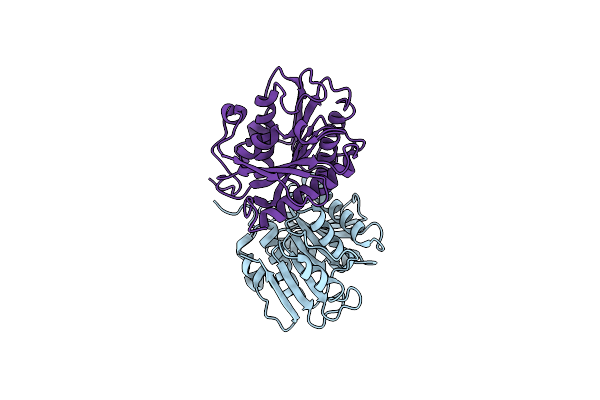 |
Crystal Structure Of Cutinase Adcut From Acidovorax Delafieldii (Pbs Depolymerase)
Organism: Acidovorax delafieldii
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2024-01-24 Classification: HYDROLASE |
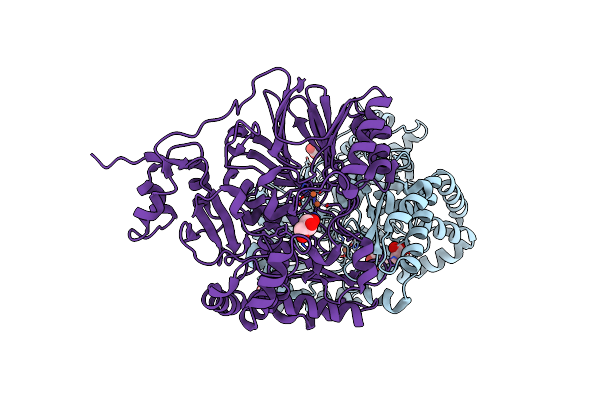 |
Organism: Chromobacterium vaccinii
Method: X-RAY DIFFRACTION Release Date: 2023-07-12 Classification: OXIDOREDUCTASE Ligands: ACT, GOL, O, FE |
 |
Organism: Rhizobacter gummiphilus
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2023-03-08 Classification: HYDROLASE Ligands: ACT |
 |
Organism: Pseudomonas saudimassiliensis
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2023-03-08 Classification: HYDROLASE Ligands: ACT |
 |
Organism: Halopseudomonas bauzanensis
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2023-03-08 Classification: HYDROLASE Ligands: SO4 |
 |
Crystal Structure Of An Alpha/Beta-Hydrolase Enzyme From Chloroflexus Sp. Ms-G (202)
Organism: Chloroflexus sp. ms-g
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2022-12-28 Classification: HYDROLASE |
 |
Crystal Structure Of An Alpha/Beta-Hydrolase Enzyme From Candidatus Kryptobacter Tengchongensis (306)
Organism: Candidatus kryptobacter tengchongensis
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: PO4 |
 |
Crystal Structure Of A Cutinase Enzyme From Marinactinospora Thermotolerans Dsm45154 (606)
Organism: Marinactinospora thermotolerans
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2022-12-28 Classification: HYDROLASE |
 |
Organism: Saccharopolyspora flava
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: PG4 |
 |
Organism: Thermobifida fusca
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: PEG |
 |
Organism: Thermobifida fusca
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: PG4 |
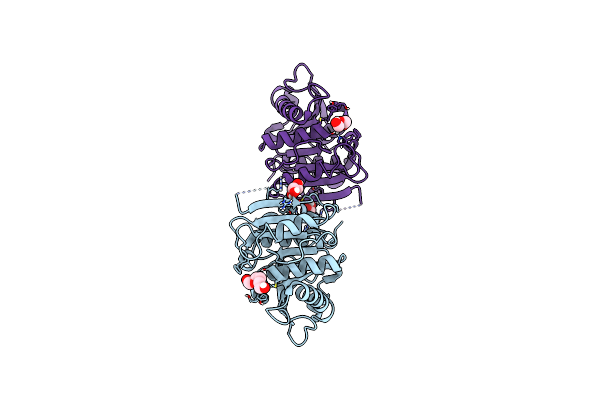 |
Organism: Thermobifida fusca yx
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: GOL, PG4, PEG |
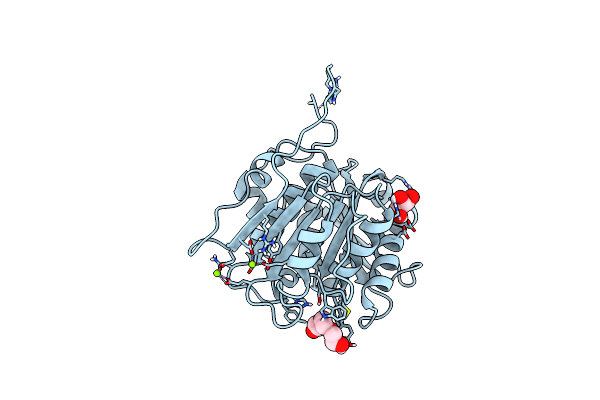 |
Crystal Structure Of A Cutinase Enzyme From Thermobifida Cellulosilytica Tb100 (711)
Organism: Thermobifida cellulosilytica tb100
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: GOL, PG4, MG |
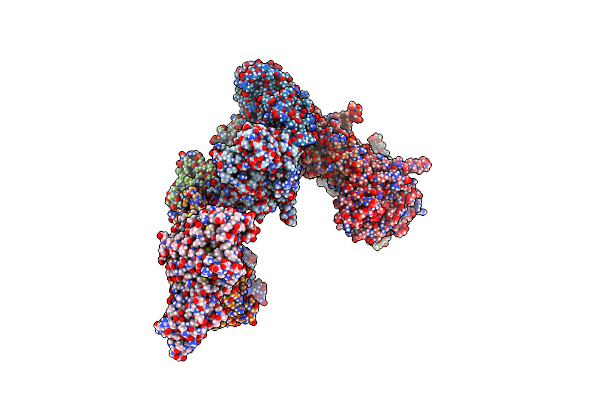 |
Organism: Rhizorhabdus wittichii rw1
Method: X-RAY DIFFRACTION Resolution:3.71 Å Release Date: 2022-06-01 Classification: OXIDOREDUCTASE Ligands: FES, FE |
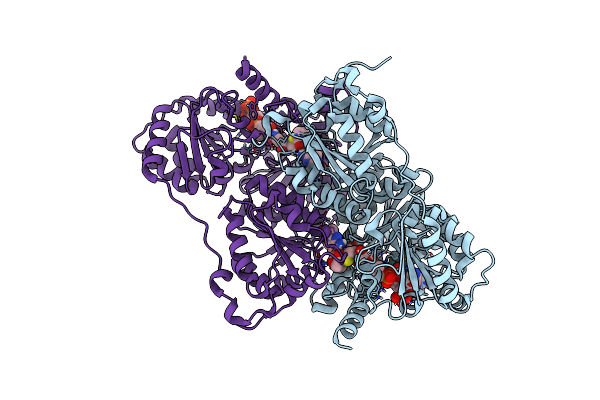 |
Actinobacterial 2-Hydroxyacyl-Coa Lyase (Achacl) Structure In Complex With Substrate 2-Hib-Coa And Inactive Cofactor 3-Deaza-Thdp
Organism: Actinomycetospora chiangmaiensis dsm 45062
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-02-02 Classification: LYASE Ligands: TPW, 3KK, MG |
 |
Actinobacterial 2-Hydroxyacyl-Coa Lyase (Achacl) Mutant E493Q Structure In Complex With Substrate 2-Hib-Coa And Inactive Cofactor 3-Deaza-Thdp
Organism: Actinomycetospora chiangmaiensis dsm 45062
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2022-02-02 Classification: LYASE Ligands: TPW, 3KK, MG |
 |
Actinobacterial 2-Hydroxyacyl-Coa Lyase (Achacl) Mutant E493A Structure In Complex With Substrate 2-Hib-Coa And Inactive Cofactor 3-Deaza-Thdp
Organism: Actinomycetospora chiangmaiensis dsm 45062
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2022-02-02 Classification: LYASE Ligands: TPW, 3KK, MG |
 |
Actinobacterial 2-Hydroxyacyl-Coa Lyase (Achacl) Structure In Complex With A Covalently Bound Reaction Intermediate As Well As Products Formyl-Coa And Acetone
Organism: Actinomycetospora chiangmaiensis dsm 45062
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2022-02-02 Classification: LYASE Ligands: TPP, SO4, ACN, FYN, MG, OXT |

