Search Count: 38
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: HYDROLASE Ligands: 4BW |
 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Resolution:2.90 Å Release Date: 2025-11-19 Classification: HYDROLASE |
 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Resolution:3.10 Å Release Date: 2025-11-19 Classification: HYDROLASE |
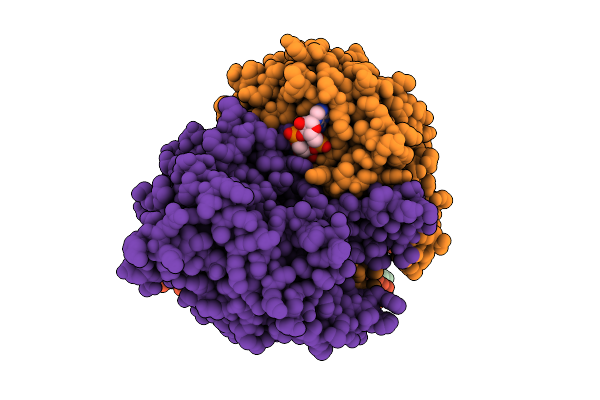 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Resolution:3.04 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: 4BW |
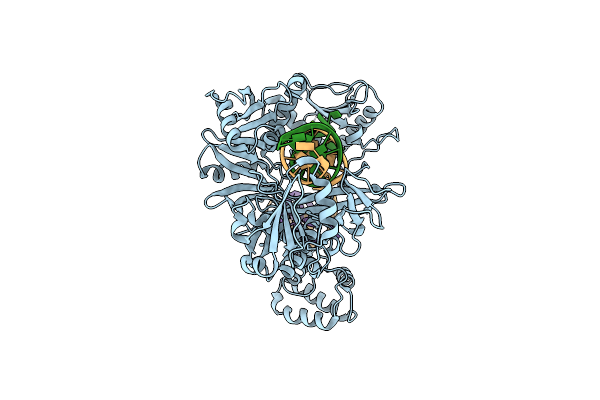 |
Organism: Lacticaseibacillus
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: DNA BINDING PROTEIN/DNA Ligands: MG |
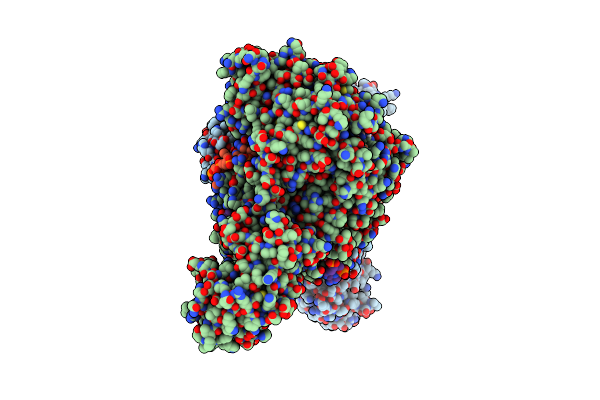 |
Organism: Lacticaseibacillus
Method: ELECTRON MICROSCOPY Resolution:2.96 Å Release Date: 2025-01-01 Classification: DNA BINDING PROTEIN/DNA Ligands: ADP |
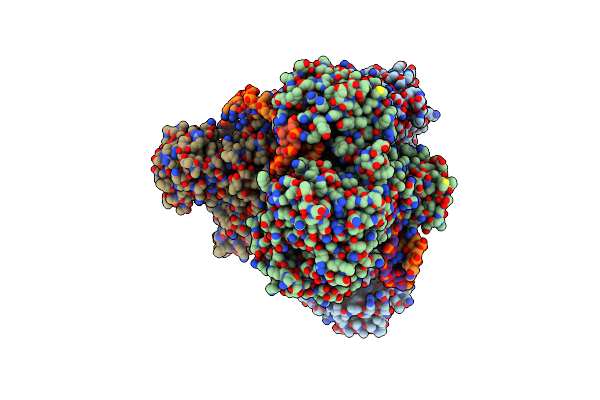 |
Organism: Lacticaseibacillus
Method: ELECTRON MICROSCOPY Resolution:3.26 Å Release Date: 2025-01-01 Classification: DNA BINDING PROTEIN/DNA Ligands: MG |
 |
The Structure Of Type Iii Crispr-Associated Deaminase In Complex Ca6 And Atp, Fully Activated
Organism: Limisphaera ngatamarikiensis
Method: ELECTRON MICROSCOPY Resolution:3.40 Å Release Date: 2024-12-25 Classification: IMMUNE SYSTEM Ligands: ATP, ZN, MG |
 |
Organism: Limisphaera ngatamarikiensis
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: IMMUNE SYSTEM Ligands: ZN, LQJ |
 |
Organism: Limisphaera ngatamarikiensis
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: IMMUNE SYSTEM Ligands: ZN, ATP |
 |
Organism: Limisphaera ngatamarikiensis
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: IMMUNE SYSTEM |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-01-24 Classification: APOPTOSIS |
 |
Organism: Homo sapiens, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-10-04 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens, Escherichia coli, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2023-10-04 Classification: SIGNALING PROTEIN Ligands: ZOB |
 |
Crystal Structure Of An Engineered Variant Of Single-Chain Penicillin G Acylase From Kluyvera Cryocrescens (Global Hydrolysis Rd3Chis)
Organism: Kluyvera cryocrescens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2021-11-17 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of An Engineered Variant Of Single-Chain Penicillin G Acylase From Kluyvera Cryocrescens (A1-Ac Rd3Chis)
Organism: Kluyvera cryocrescens
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2021-11-17 Classification: HYDROLASE Ligands: CA, PMS |
 |
Crystal Structure Of The Sars-Cov-2 (Covid-19) Main Protease In Complex With Z-Vad-Fmk
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2021-06-23 Classification: VIRAL PROTEIN Ligands: DMS, GOL |
 |
Crystal Structure Of The Sars-Cov-2 (Covid-19) Main Protease In Complex With Mg132
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2021-06-23 Classification: VIRAL PROTEIN Ligands: DMS, ALD, GOL |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-10-17 Classification: HYDROLASE Ligands: DGT, MG |

