Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: NUCLEAR PROTEIN Ligands: A1MAM, EDO, ACT |
 |
Structure-Based Characterization And Improvement Of An Enzymatic Activity Of Acremonium Alcalophilum Feruloyl Esterase
Organism: Sodiomyces alcalophilus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-04-17 Classification: HYDROLASE Ligands: PEG, MG |
 |
Structure-Based Characterization And Improvement Of An Enzymatic Activity Of Acremonium Alcalophilum Feruloyl Esterase
Organism: Sodiomyces alcalophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-04-17 Classification: HYDROLASE Ligands: FER, MG |
 |
Structure Insight Into Substrate Recognition And Catalysis By Feruloyl Esterase From Aspergillus Sydowii
Organism: Aspergillus sydowii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-10-25 Classification: HYDROLASE Ligands: NAG, ACT |
 |
Structure Insight Into Substrate Recognition And Catalysis By Feruloyl Esterase From Aspergillus Sydowii
Organism: Aspergillus sydowii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-10-25 Classification: HYDROLASE Ligands: NAG, FER, ACT, EDO |
 |
Structure Insight Into Substrate Recognition And Catalysis By Feruloyl Esterase From Aspergillus Sydowii
Organism: Aspergillus sydowii
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-10-25 Classification: HYDROLASE Ligands: SXX, NAG, EDO, ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-09-21 Classification: TRANSCRIPTION Ligands: ZN, CA, NA, KIZ, PG4, PEG, KZF |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2022-09-21 Classification: TRANSCRIPTION Ligands: ZN, CA, NA, PGE, EDO, KJ6, 2OP, PG4, PEG, NHE, DMS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2022-09-21 Classification: TRANSCRIPTION Ligands: ZN, CA, NA, EDO, KJN, NHE, PG4, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2022-09-21 Classification: TRANSCRIPTION Ligands: ZN, CA, NA, PEG, EDO, SHH, KK0, KZF, PG4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2022-09-21 Classification: TRANSCRIPTION Ligands: ZN, CA, NA, PEG, PGE, KJF, NHE, PG4, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-09-21 Classification: TRANSCRIPTION Ligands: EDO, PG4, PEG, ZN, CA, NA, KKI, NHE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2022-09-21 Classification: TRANSCRIPTION Ligands: PEG, PG4, EDO, ZN, CA, NA, KKW, KZF |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-09-21 Classification: TRANSCRIPTION Ligands: ZN, CA, NA, PEG, KRU, NHE, PG4, EDO |
 |
Organism: Ceriporiopsis subvermispora (strain b)
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2022-05-04 Classification: OXIDOREDUCTASE Ligands: NAG, CU, TRS, EDO, XYS, CA |
 |
Crystal Structure Of An Apo Form Of The Glutathione S-Transferase, Csgst83044, Of Ceriporiopsis Subvermispora
Organism: Ceriporiopsis subvermispora (strain b)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-05-15 Classification: TRANSFERASE Ligands: EDO, CA, SO4 |
 |
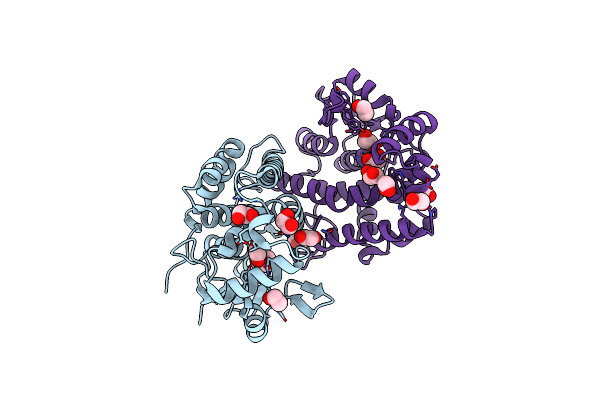
Crystal Structure Of The Glutathione S-Transferase, Csgst83044, Of Ceriporiopsis Subvermispora In Complex With Glutathione
Organism: Ceriporiopsis subvermispora (strain b)
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2019-05-15 Classification: TRANSFERASE Ligands: GSH |
 |
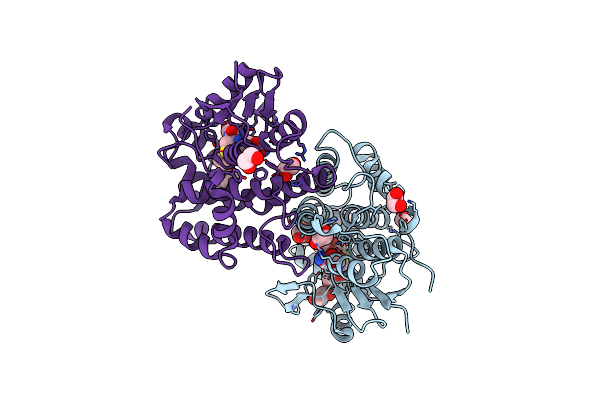
Crystal Structure Of An Apo Form Of The Glutathione S-Transferase, Csgst63524, Of Ceriporiopsis Subvermispora
Organism: Ceriporiopsis subvermispora (strain b)
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2019-02-27 Classification: TRANSFERASE Ligands: EDO |
 |
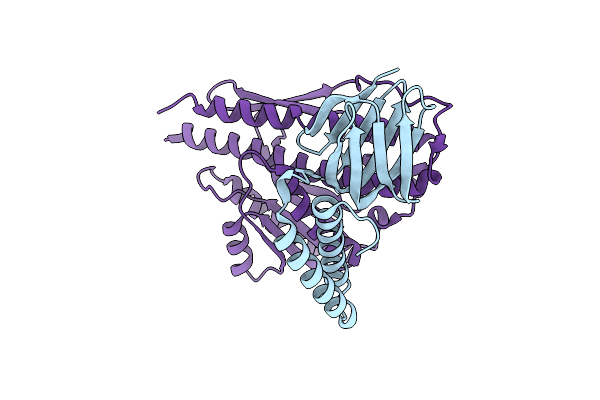
Crystal Structure Of The Glutathione S-Transferase, Csgst63524, Of Ceriporiopsis Subvermispora In Complex With Glutathione
Organism: Ceriporiopsis subvermispora (strain b)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-02-27 Classification: TRANSFERASE Ligands: GSH, EDO |
 |
Crystal Structure Of The Gamma - Epsilon Complex Of Photosynthetic Cyanobacterial F1-Atpase
Organism: Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2018-09-26 Classification: HYDROLASE |