Search Count: 49
 |
A Ternary Complex Of Plant Adenosine Kinase 1 From Moss Physcomitrella Patens (Ppadk1) With Adenosine And Adp
Organism: Physcomitrium patens
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2025-04-09 Classification: TRANSFERASE Ligands: ADP, ADN, NA, EDO |
 |
Crystal Structure Of Zea Mays Adenosine Kinase 3 (Zmadk3) In Complex With Ap5A
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2025-01-29 Classification: TRANSFERASE Ligands: AP5, EDO, PEG, CL |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2025-01-01 Classification: TRANSFERASE Ligands: SO4, ACT, GOL |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2025-01-01 Classification: TRANSFERASE Ligands: ACP, ACT, GOL |
 |
Structure Of Aldh5F1 From Moss Physcomitrium Patens In Complex With Nad+ In The Contracted Conformation
Organism: Physcomitrium patens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2024-03-27 Classification: OXIDOREDUCTASE Ligands: NAD, P33, PEG, EDO, SO4 |
 |
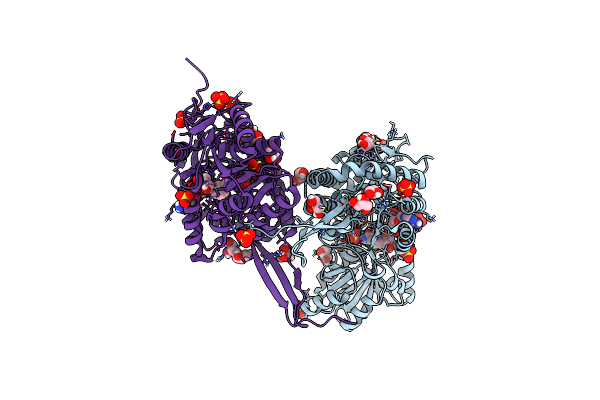
Structure Of The Apoform Of Aldehyde Dehydrogenase 5F1 (Aldh5F1) From The Moss Physcomitrium Patens
Organism: Physcomitrium patens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2024-03-27 Classification: OXIDOREDUCTASE Ligands: P33, PGE, 1PE, EDO, SO4 |
 |
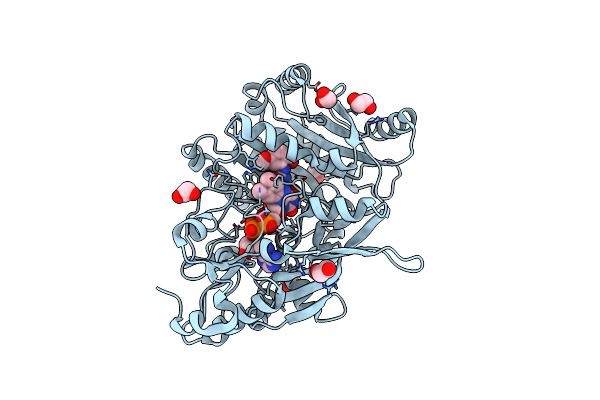
Structure Of The Aldehyde Dehydrogenase 5F1 (Aldh5F1) From The Moss Physcomitrium Patens In Complex With Nad In An Extended Conformation
Organism: Physcomitrium patens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2024-03-27 Classification: OXIDOREDUCTASE Ligands: EDO, GOL, NAD, SO4 |
 |
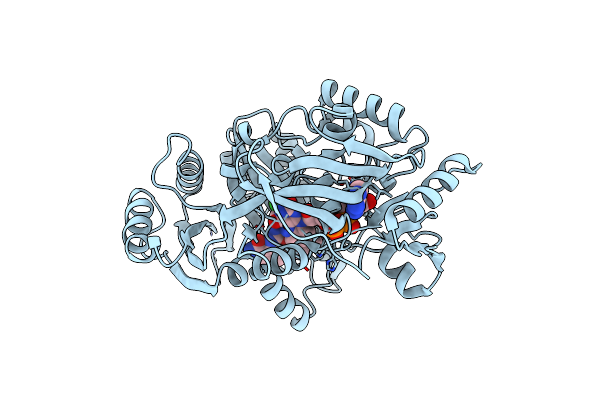
Crystal Structure Of Maize Cko/Ckx8 In Complex With Urea-Derived Inhibitor 2-[(3,5-Dichlorophenyl)Carbamoylamino]-4-Methoxy-Benzamide
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-02-28 Classification: FLAVOPROTEIN Ligands: EDO, GOL, FAD, UZ3 |
 |
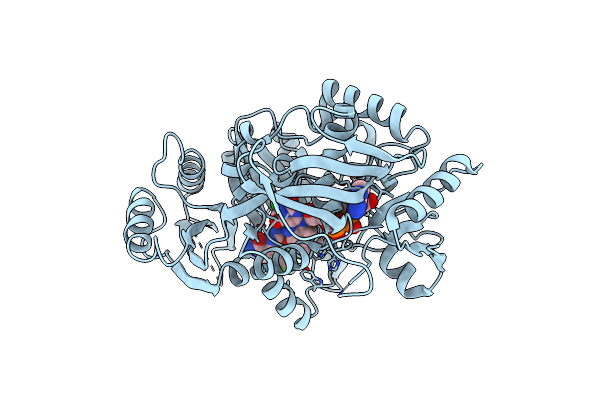
Crystal Structure Of Maize Cytokinin Oxidase/Dehydrogenase 4 (Cko/Ckx4) In Complex With Inhibitor 2-[(3,5-Dichlorophenyl)Carbamoylamino]Benzamide
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-02-28 Classification: FLAVOPROTEIN Ligands: FAD, UZX |
 |
Crystal Structure Of Maize Cytokinin Oxidase/Dehydrogenase 4 (Cko/Ckx4) In Complex With Inhibitor 2-[(3,5-Dichlorophenyl)Carbamoylamino]-4-(Trifluoromethoxy)Benzamide
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-02-28 Classification: FLAVOPROTEIN Ligands: FAD, V2F |
 |
Crystal Structure Of Maize Cytokinin Oxidase/Dehydrogenase 4 (Cko/Ckx4) In Complex With Inhibitor 2-[(3,5-Dichlorophenyl)Carbamoylamino]-4-Methoxy-Benzamide
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-02-28 Classification: FLAVOPROTEIN Ligands: FAD, UZ3, GOL, EDO |
 |
Crystal Structure Of Maize Cytokinin Oxidase/Dehydrogenase 4 (Cko/Ckx4) In Complex With Inhibitor 2-[[3,5-Dichloro-2-(2-Hydroxyethyl)Phenyl]Carbamoylamino]-4-(Trifluoromethoxy)Benzamide
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-02-28 Classification: FLAVOPROTEIN Ligands: V1X, FAD |
 |
Crystal Structure Of Maize Cko/Ckx8 In Complex With Urea-Derived Inhibitor 2-[(3,5-Dichlorophenyl)Carbamoylamino]Benzamide
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2024-02-21 Classification: FLAVOPROTEIN Ligands: FAD, UZX, GOL, EDO |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2022-01-12 Classification: HYDROLASE Ligands: CA, GOL, EDO, CL, PEG |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-01-12 Classification: HYDROLASE Ligands: CA, IMH, DMS, EDO, PEG, PGE |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-01-12 Classification: HYDROLASE Ligands: CA, RIB, EDO, PGE, PEG |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-01-12 Classification: HYDROLASE Ligands: CA, ADE, PEG, EDO |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-01-12 Classification: HYDROLASE Ligands: CA, IMH, EDO, PGE |
 |
Crystal Structure Of Zmcko4A In Complex With Inhibitor 1-[2-(2-Hydroxy-Ethyl)-Phenyl]-3-(3-Trifluoromethoxy-Phenyl)-Urea
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-10-14 Classification: FLAVOPROTEIN Ligands: FAD, OJ2, EDO, DMS |
 |
Crystal Structure Of Zmcko4A In Complex With Inhibitor 1-(3-Chloro-5-Trifluoromethoxy-Phenyl)-3-[2-(2-Hydroxy-Ethyl)-Phenyl]-Urea
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-10-14 Classification: FLAVOPROTEIN Ligands: EDO, FAD, OHZ, DMS |

