Search Count: 17
 |
Organism: Tomato spotted wilt virus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-03-22 Classification: VIRAL PROTEIN |
 |
Organism: Tomato spotted wilt virus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2017-03-22 Classification: VIRAL PROTEIN/RNA |
 |
Organism: Tomato spotted wilt virus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2017-03-22 Classification: Viral protein/DNA |
 |
Crystal Structure Of Ruminococcus Albus 4-O-Beta-D-Mannosyl-D-Glucose Phosphorylase (Ramp1)
Organism: Ruminococcus albus (strain atcc 27210 / dsm 20455 / jcm 14654 / ncdo 2250 / 7)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-03-23 Classification: TRANSFERASE |
 |
Crystal Structure Of Ruminococcus Albus 4-O-Beta-D-Mannosyl-D-Glucose Phosphorylase (Ramp1) In Complexes With Sulfate And 4-O-Beta-D-Mannosyl-D-Glucose
Organism: Ruminococcus albus (strain atcc 27210 / dsm 20455 / jcm 14654 / ncdo 2250 / 7)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-03-23 Classification: TRANSFERASE Ligands: SO4 |
 |
Crystal Structure Of Ruminococcus Albus Beta-(1,4)-Mannooligosaccharide Phosphorylase (Ramp2) In Complexes With Phosphate
Organism: Ruminococcus albus (strain atcc 27210 / dsm 20455 / jcm 14654 / ncdo 2250 / 7)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-03-23 Classification: TRANSFERASE Ligands: PO4 |
 |
Crystal Structure Of Ruminococcus Albus Beta-(1,4)-Mannooligosaccharide Phosphorylase (Ramp2) In Complexes With Phosphate And Beta-(1,4)-Mannobiose
Organism: Ruminococcus albus (strain atcc 27210 / dsm 20455 / jcm 14654 / ncdo 2250 / 7)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-03-23 Classification: TRANSFERASE Ligands: PO4 |
 |
Organism: Halomonas sp. h11
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2015-06-10 Classification: HYDROLASE Ligands: MG, GOL, PRU |
 |
Organism: Halomonas sp. h11
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2015-06-10 Classification: HYDROLASE Ligands: MG, GOL, BGC |
 |
Crystal Structure Of Alpha-Glucosidase Mutant D202N In Complex With Glucose And Glycerol
Organism: Halomonas sp. h11
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2015-06-10 Classification: HYDROLASE Ligands: MG, GOL, BGC |
 |
Crystal Structure Of Alpha-Glucosidase Mutant E271Q In Complex With Maltose
Organism: Halomonas sp. h11
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-06-10 Classification: HYDROLASE Ligands: MG, GOL |
 |
Organism: Emericella nidulans (strain fgsc a4 / atcc 38163 / cbs 112.46 / nrrl 194 / m139)
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2015-06-10 Classification: TRANSLATION |
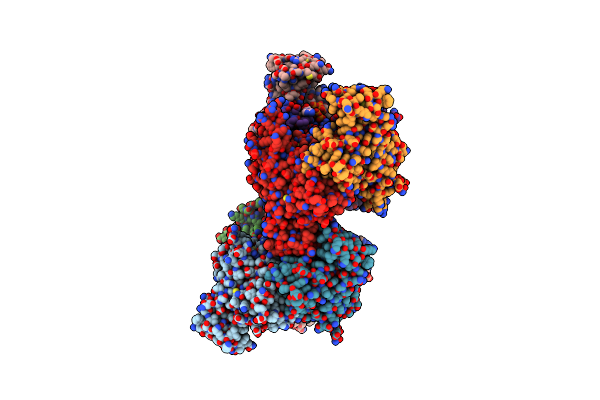 |
Crystal Structure Of C. Albicans Trna(His) Guanylyltransferase (Thg1) With Atp
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2013-12-18 Classification: TRANSFERASE Ligands: ATP, MG |
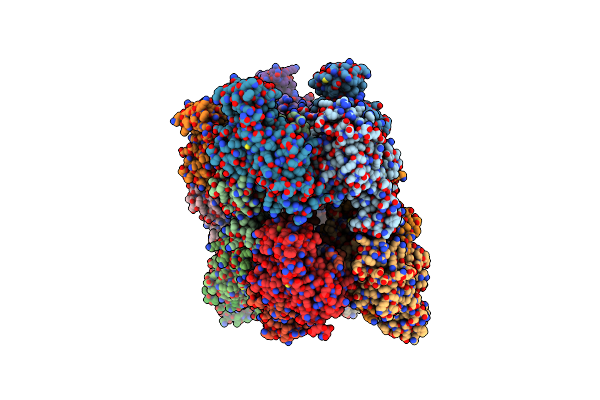 |
Crystal Structure Of C. Albicans Trna(His) Guanylyltransferase (Thg1) With Gtp
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2013-12-18 Classification: TRANSFERASE Ligands: GTP, MG |
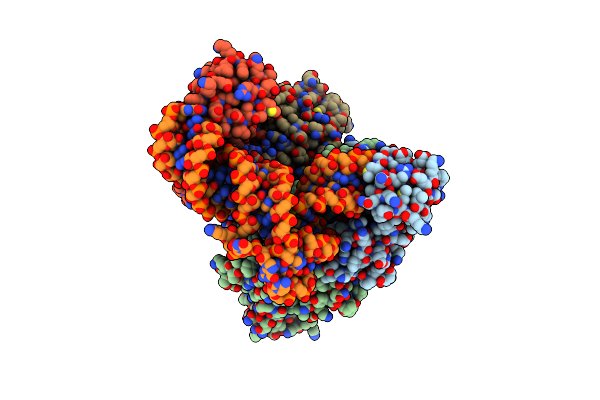 |
Crystal Structure Of C. Albicans Trna(His) Guanylyltransferase (Thg1) With A G-1 Deleted Trna(His)
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:4.18 Å Release Date: 2013-12-18 Classification: TRANSFERASE/RNA |
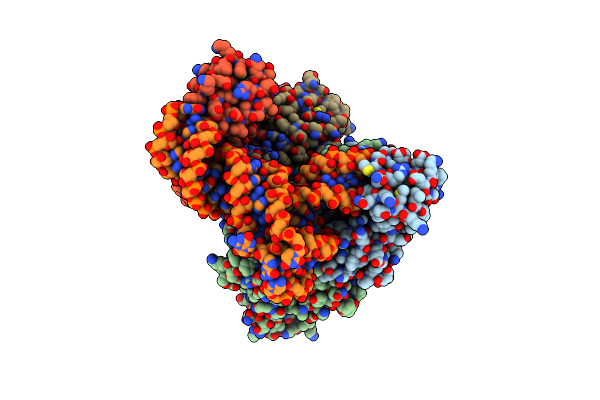 |
Crystal Structure Of C. Albicans Trna(His) Guanylyltransferase (Thg1) With A Trna(Phe)(Gug)
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:3.64 Å Release Date: 2013-12-18 Classification: TRANSFERASE/RNA |
 |
Crystal Structure Of Pilz Domain Of Cesa From Cellulose Synthesizing Bacterium
Organism: Gluconacetobacter xylinus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-04-03 Classification: TRANSFERASE |

