Search Count: 10
 |
Crystal Structure Of The Manihot Esculenta Hydroxynitrile Lyase (Mehnl) 3Kp (K176P, K199P, K224P) Triple Mutant
Organism: Manihot esculenta
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2016-03-02 Classification: LYASE |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-10-14 Classification: HYDROLASE Ligands: SCN |
 |
Crystal Structure Of The Manihot Esculenta Hydroxynitrile Lyase (Mehnl) K176P Mutant
Organism: Manihot esculenta
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-06-20 Classification: LYASE Ligands: GOL |
 |
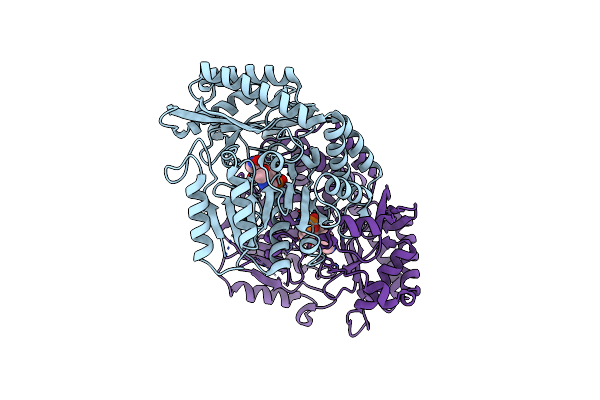
The Crystal Structure Of Alpha-Amino-Epsilon-Caprolactam Racemase From Achromobacter Obae Complexed With Epsilon Caprolactam
Organism: Achromobacter obae
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2009-07-28 Classification: ISOMERASE Ligands: PLP, ICC |
 |
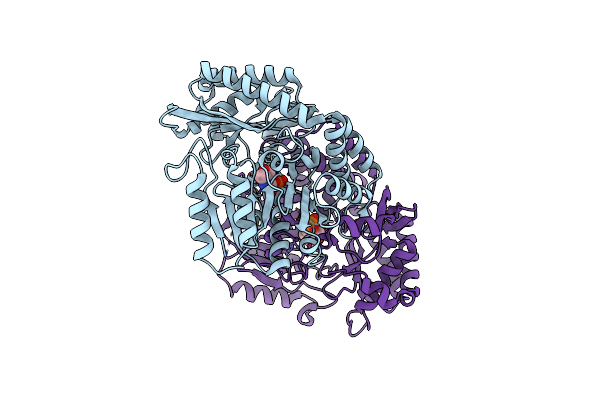
The Crystal Structure Of Alpha-Amino-Epsilon-Caprolactam Racemase From Achromobacter Obae Complexed With Epsilon Caprolactam (Different Binding Mode)
Organism: Achromobacter obae
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2009-02-17 Classification: ISOMERASE Ligands: ICC, PLP |
 |
The Crystal Structure Of Alpha-Amino-Epsilon-Caprolactam Racemase From Achromobacter Obae
Organism: Achromobacter obae
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2009-02-17 Classification: ISOMERASE Ligands: PLP |
 |
The Crystal Structure Of D-Amino Acid Amidase From Ochrobactrum Anthropi Sv3 Complexed With L-Phenylalanine
Organism: Ochrobactrum anthropi
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2007-03-06 Classification: HYDROLASE Ligands: BA, PHE |
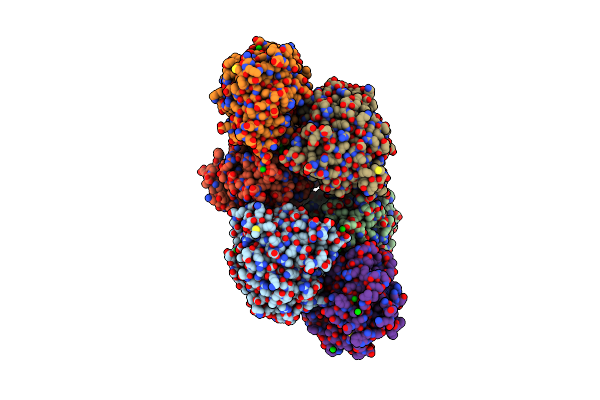 |
The Crystal Structure Of D-Amino Acid Amidase From Ochrobactrum Anthropi Sv3 Complexed With L-Phenylalanine Amide
Organism: Ochrobactrum anthropi
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-03-06 Classification: HYDROLASE Ligands: BA, NFA |
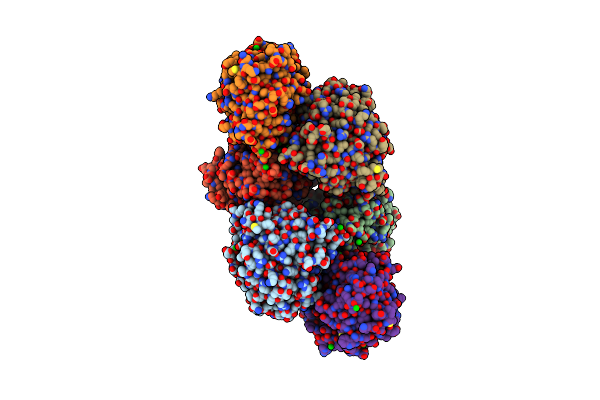 |
The Crystal Structutre Of D-Amino Acid Amidase From Ochrobactrum Anthropi Sv3
Organism: Ochrobactrum anthropi
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2006-07-04 Classification: HYDROLASE Ligands: BA |
 |
The Crystal Structure Of D-Amino Acid Amidase From Ochrobactrum Anthropi Sv3 Complexed With D-Phenylalanine
Organism: Ochrobactrum anthropi
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2006-05-09 Classification: HYDROLASE Ligands: BA, DPN |

