Search Count: 23
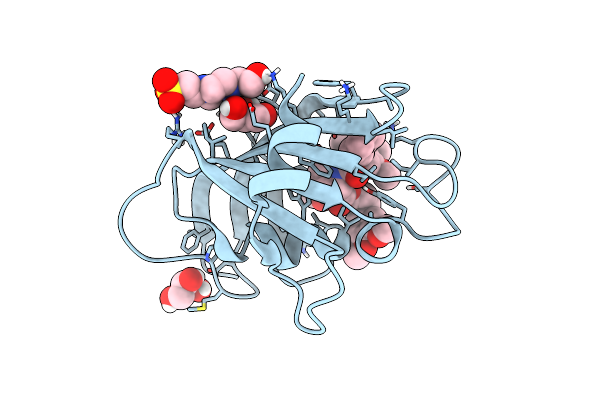 |
Structure Of The Fk1 Domain Of The Fkbp51 G64S Variant In Complex With Safit1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2022-11-16 Classification: ISOMERASE Ligands: GY1, EPE, GOL |
 |
Structure Of The Fk1 Domain Of The Fkbp51 G64S Variant In Complex With (2R,5S,12R)-12-Cyclohexyl-2-[2-(3,4-Dimethoxyphenyl)Ethyl]-15,15,16-Trimethyl-3,19-Dioxa-10,13,16-Triazatricyclo[18.3.1.0^5,^10]Tetracosa-1(24),20,22-Triene-4,11,14,17-Tetrone
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2022-11-16 Classification: ISOMERASE Ligands: S8N |
 |
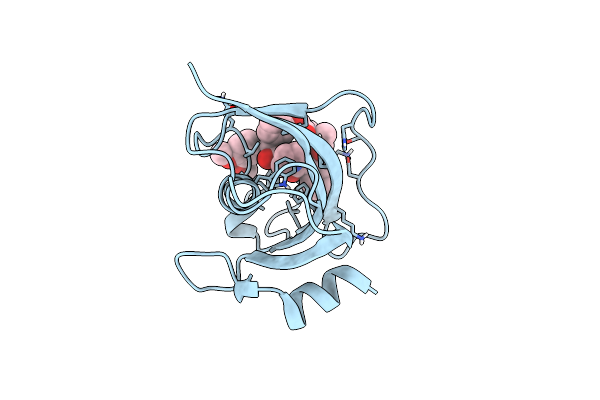
Structure Of The Fk1 Domain Of The Fkbp51 G64S Variant In Complex With (1S,5S,6R)-10-((3,5-Dichlorophenyl)Sulfonyl)-5-(Hydroxymethyl)-3-(Pyridin-2-Ylmethyl)-3,10-Diazabicyclo[4.3.1]Decan-2-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2022-11-16 Classification: ISOMERASE Ligands: 9QN |
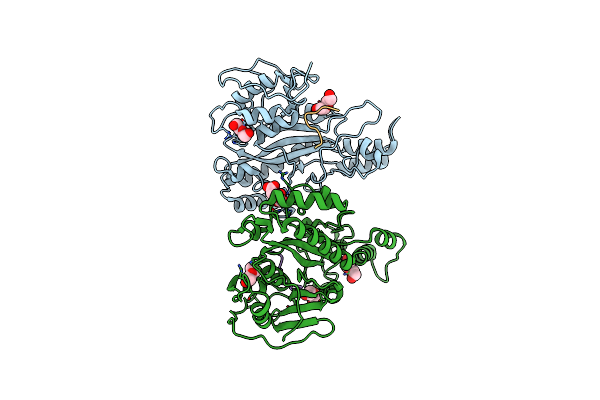 |
Structure Of A Glutamine Donor Mimicking Inhibitory Peptide Shaped By The Catalytic Cleft Of Microbial Transglutaminase
Organism: Streptomyces mobaraensis, Streptomyces mobaraensis nbrc 13819 = dsm 40847
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2018-10-24 Classification: TRANSFERASE Ligands: FLC, PEG |
 |
Organism: Streptomyces mobaraensis, Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-09-12 Classification: ANTIMICROBIAL PROTEIN |
 |
Structure Of The Dispase Autolysis Inducing Protein From Streptomyces Mobaraensis
Organism: Streptomyces mobaraensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-08-10 Classification: SIGNALING PROTEIN Ligands: GOL, CA |
 |
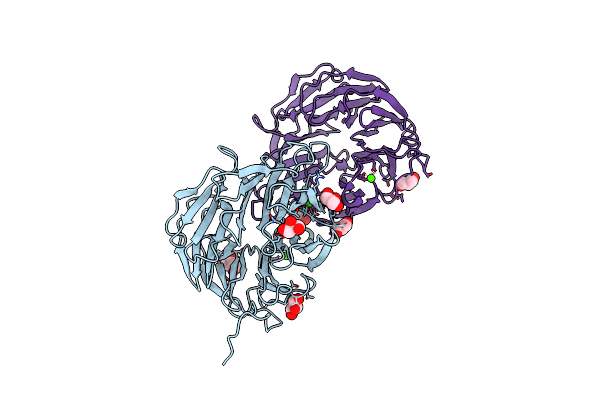
Cationic Trypsin In Complex With Mutated Spinacia Oleracea Trypsin Inhibitor Iii (Soti-Iii) (F14A)
Organism: Spinacia oleracea, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-01-09 Classification: HYDROLASE/INHIBITOR Ligands: CA, 1PE |
 |
Cationic Trypsin In Complex With The Spinacia Oleracea Trypsin Inhibitor Iii (Soti-Iii)
Organism: Spinacia oleracea, Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-01-09 Classification: HYDROLASE/INHIBITOR Ligands: GOL, CL, IMD, CA, MES |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2013-01-09 Classification: IMMUNE SYSTEM Ligands: CL |
 |
Co-Complex Structure Of Bovine Trypsin With A Modified Bowman-Birk Inhibitor (Pta)Sfti-1(1,14), That Was 1,4-Disubstituted With A 1,2,3- Trizol To Mimic A Trans Amide Bond
Organism: Synthetic construct, Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2012-03-07 Classification: HYDROLASE/INHIBITOR Ligands: CA, DMF, GOL, SO4 |
 |
Co-Complex Structure Of Bovine Trypsin With A Modified Bowman-Birk Inhibitor (Ica)Sfti-1(1,14), That Was 1,5-Disubstituted With 1,2,3- Trizol To Mimic A Cis Amide Bond
Organism: Helianthus annuus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2012-03-07 Classification: HYDROLASE/INHIBITOR Ligands: CA, DMF, GOL, SO4 |
 |
 |
Solution Structure Of A Linear Analog Of The Cyclic Squash Trypsin Inhibitor Mcoti-Ii
Organism: Momordica cochinchinensis
Method: SOLUTION NMR Release Date: 2007-10-02 Classification: PLANT PROTEIN |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-08-14 Classification: OXIDOREDUCTASE Ligands: PEG, CL, FAD |
 |
Crystal Structure Of The Electron Transfer Complex Rubredoxin - Rubredoxin Reductase From Pseudomonas Aeruginosa.
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2007-08-14 Classification: OXIDOREDUCTASE Ligands: FAD, FE |
 |
Crystal Structure Of Sdsa1, An Alkylsulfatase From Pseudomonas Aeruginosa, In Complex With 1-Decane-Sulfonic-Acid.
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-04-26 Classification: HYDROLASE Ligands: 1DB, PEG, IPA, ZN |
 |
Crystal Structure Of Sdsa1, An Alkylsulfatase From Pseudomonas Aeruginosa, In Complex With 1-Dodecanol
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2006-04-26 Classification: HYDROLASE Ligands: 1DO, PEG, ZN |
 |
Crystal Structure Of Sdsa1, An Alkylsulfatase From Pseudomonas Aeruginosa, In Complex With Sulfate
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2006-04-26 Classification: HYDROLASE Ligands: SO4, ZN |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2006-04-26 Classification: HYDROLASE Ligands: ZN |
 |
Inhibitor Cystine Knot Protein Mcoeeti Fused To The Catalytically Inactive Barnase Mutant H102A
Organism: Bacillus amyloliquefaciens, Momordica cochinchinensis, Ecballium elaterium
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2005-11-21 Classification: FUSION PROTEIN Ligands: 2PE, GOL, EDO, FMT, SO4, UNX, MES |

