Search Count: 19
 |
Organism: Thermochaetoides thermophila
Method: ELECTRON MICROSCOPY Release Date: 2023-08-02 Classification: DNA BINDING PROTEIN Ligands: ADP, MG, ATP |
 |
Organism: Thermochaetoides thermophila, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-07-26 Classification: DNA BINDING PROTEIN Ligands: ADP, ALF, MG, ATP |
 |
Organism: Thermochaetoides thermophila, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-07-26 Classification: DNA BINDING PROTEIN Ligands: ADP, MG, ALF |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-07-26 Classification: DNA BINDING PROTEIN |
 |
Organism: Thermochaetoides thermophila
Method: ELECTRON MICROSCOPY Release Date: 2023-07-26 Classification: DNA BINDING PROTEIN Ligands: ATP, MG |
 |
Cryoem Structure Ino80Core Hexasome Complex Arp5 Grappler Refinement State1
Organism: Thermochaetoides thermophila
Method: ELECTRON MICROSCOPY Release Date: 2023-07-26 Classification: DNA BINDING PROTEIN |
 |
Organism: Thermochaetoides thermophila, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-07-26 Classification: DNA BINDING PROTEIN Ligands: ADP, MG, ALF, ATP |
 |
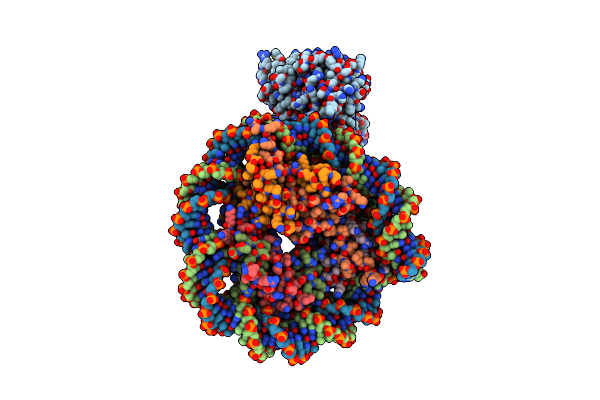
Organism: Thermochaetoides thermophila
Method: ELECTRON MICROSCOPY Release Date: 2023-07-26 Classification: DNA BINDING PROTEIN Ligands: ADP, ATP, MG |
 |
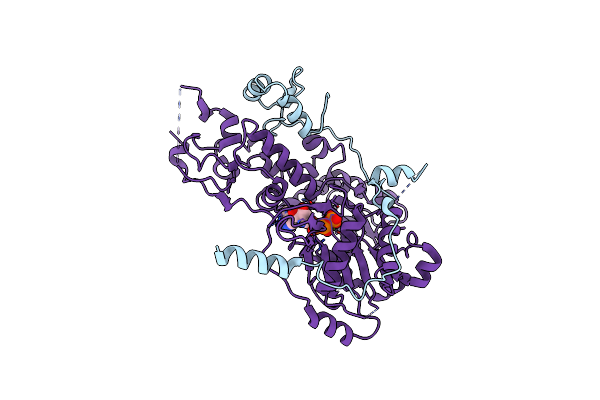
Cryoem Structure Ino80Core Hexasome Complex Atpase-Hexasome Refinement State 2
Organism: Thermochaetoides thermophila, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-07-26 Classification: DNA BINDING PROTEIN Ligands: ADP, MG, ALF |
 |
Organism: Thermochaetoides thermophila
Method: ELECTRON MICROSCOPY Release Date: 2023-07-26 Classification: DNA BINDING PROTEIN Ligands: ATP, MG |
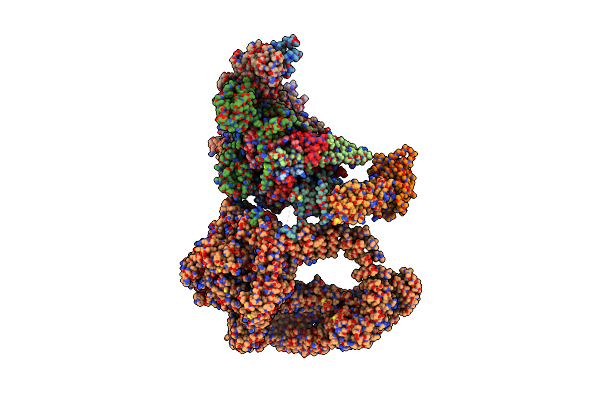 |
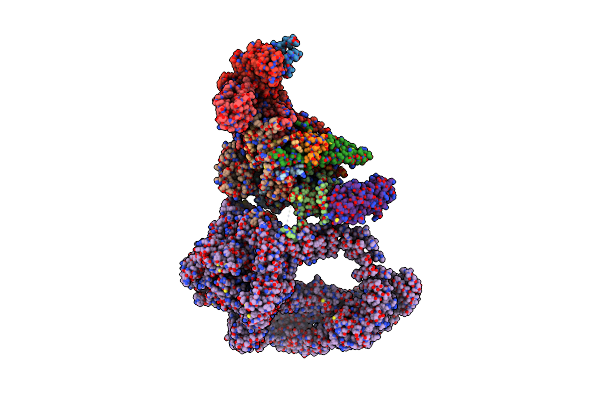
Organism: Komagataella phaffii (strain gs115 / atcc 20864), Homo sapiens, Komagataella phaffii gs115
Method: ELECTRON MICROSCOPY Release Date: 2020-02-12 Classification: TRANSCRIPTION |
 |
Organism: Komagataella phaffii (strain gs115 / atcc 20864), Komagataella phaffii gs115
Method: ELECTRON MICROSCOPY Release Date: 2020-01-29 Classification: TRANSCRIPTION |
 |
Organism: Komagataella phaffii gs115, Komagataella phaffii (strain gs115 / atcc 20864)
Method: ELECTRON MICROSCOPY Release Date: 2018-11-21 Classification: TRANSCRIPTION |
 |
Structure Of C-Terminal Domain From S. Cerevisiae Pat1 Decapping Activator Bound To Dcp2 Hlm2 Peptide (Region 435-451)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-08-16 Classification: RNA BINDING PROTEIN |
 |
Structure Of C-Terminal Domain From S. Cerevisiae Pat1 Decapping Activator Bound To Dcp2 Hlm3 Peptide (Region 484-500)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-08-16 Classification: RNA BINDING PROTEIN Ligands: MG, CL, EDO |
 |
Structure Of C-Terminal Domain From S. Cerevisiae Pat1 Decapping Activator Bound To Dcp2 Hlm10 Peptide (Region 954-970)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2017-08-16 Classification: RNA BINDING PROTEIN Ligands: EDO, MG, TRS |
 |
Organism: Komagataella pastoris
Method: ELECTRON MICROSCOPY Resolution:5.70 Å Release Date: 2017-08-02 Classification: TRANSCRIPTION |
 |
Structure Of C-Terminal Domain From S. Cerevisiae Pat1 Decapping Activator (Space Group : P21)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-10-08 Classification: RNA BINDING PROTEIN Ligands: MES, EDO |
 |
Structure Of C-Terminal Domain From S. Cerevisiae Pat1 Decapping Activator (Space Group : P212121)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2014-10-08 Classification: CELL CYCLE Ligands: MG, EDO, CL |

