Search Count: 26
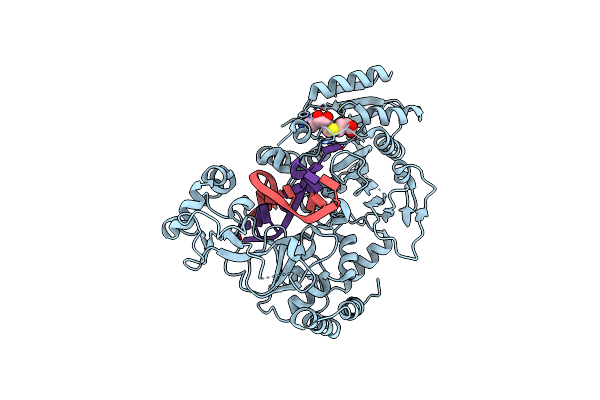 |
Adenine-Specific Dna Methyltransferase M.Bseci Complexed With Adohcy And Cognate Unmethylated Dna Duplex
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Release Date: 2023-01-25 Classification: TRANSFERASE Ligands: SAH |
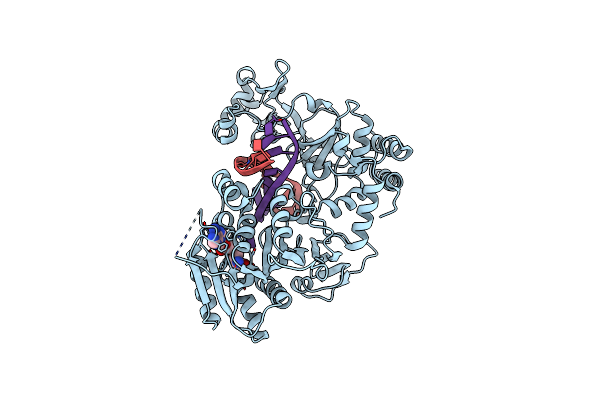 |
Adenine-Specific Dna Methyltransferase M.Bseci Complexed With Adohcy And Cognate Hemimethylated Dna Duplex
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Release Date: 2023-01-25 Classification: TRANSFERASE Ligands: SAH |
 |
Adenine-Specific Dna Methyltransferase M.Bseci Complexed With Adohcy And Cognate Fully Methylated Dna Duplex
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Release Date: 2023-01-25 Classification: TRANSFERASE Ligands: SAH |
 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-01-25 Classification: TRANSFERASE |
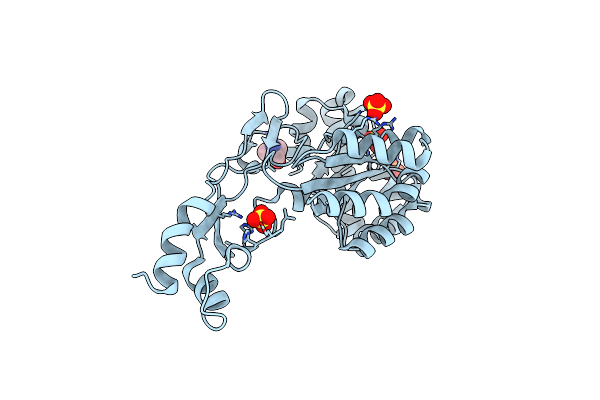 |
Crystal Structure Of Wt Ba3943, A Ce4 Family Pseudoenzyme From Bacillus Anthracis
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2021-05-19 Classification: UNKNOWN FUNCTION Ligands: SO4, TRS |
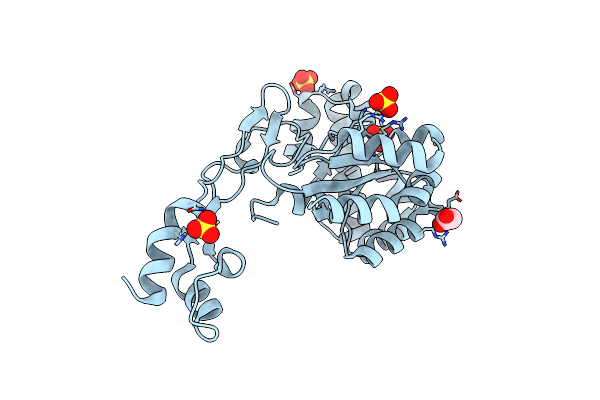 |
Crystal Structure Of A Ba3943 Mutant,A Ce4 Family Pseudoenzyme With Restored Enzymatic Activity.
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2019-10-09 Classification: UNKNOWN FUNCTION Ligands: SO4, ACT |
 |
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-10-09 Classification: UNKNOWN FUNCTION Ligands: SO4, TRS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.93 Å Release Date: 2019-04-10 Classification: OXIDOREDUCTASE Ligands: PO4, NA, CL |
 |
Crystal Structure Of The Bc1960 Peptidoglycan N-Acetylglucosamine Deacetylase In Complex With 4-Naphthalen-1-Yl-~{N}-Oxidanyl-Benzamide
Organism: Bacillus cereus (strain atcc 14579 / dsm 31 / jcm 2152 / nbrc 15305 / ncimb 9373 / nrrl b-3711)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-06-20 Classification: HYDROLASE Ligands: SO4, 5YA, PE3, PEG, PG0, NA, CL, ZN, DMS |
 |
Crystal Structure Of The Bc1960 Peptidoglycan N-Acetylglucosamine Deacetylase From Bacillus Cereus
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2014-06-04 Classification: HYDROLASE Ligands: SO4, ACT |
 |
Crystal Structure Of The Rop Protein Mutant D30P/A31G At Resolution 1.4 Resolution.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2013-02-13 Classification: RNA BINDING PROTEIN |
 |
Crystal Structure Of Bc0361, A Polysaccharide Deacetylase From Bacillus Cereus
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-10-10 Classification: HYDROLASE Ligands: ZN, ACT |
 |
Organism: Proteus hauseri
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2010-04-14 Classification: HYDROLASE Ligands: SO4, MPD, TRS |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-07-17 Classification: HYDROLASE Ligands: ZN, ACT |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2004-09-28 Classification: TRANSCRIPTION |
 |
Crystal Structure Of Chloramphenicol Acetyltransferase I Complexed With Fusidic Acid At 2.18 A Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2004-08-03 Classification: TRANSFERASE Ligands: CA, FUA |
 |
Crystal Structure Of E.Coli Chloramphenicol Acetyltransferase Type I At 2.5 Angstrom Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2004-06-01 Classification: TRANSFERASE |
 |
Crystal Structure Of The C-Terminal Domain Of The Hrcqb Protein From Pseudomonas Syringae Pv. Phaseolicola
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2003-12-04 Classification: STRUCTURAL PROTEIN |
 |
Structural And Biochemical Characterization Of A New Magnesium Ion Binding Site Near Tyr94 In The Restriction Endonuclease Pvuii
Organism: Proteus vulgaris
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2003-08-07 Classification: ENDONUCLEASE Ligands: MG |
 |
Organism: Proteus vulgaris
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2003-02-04 Classification: HYDROLASE |

