Search Count: 27
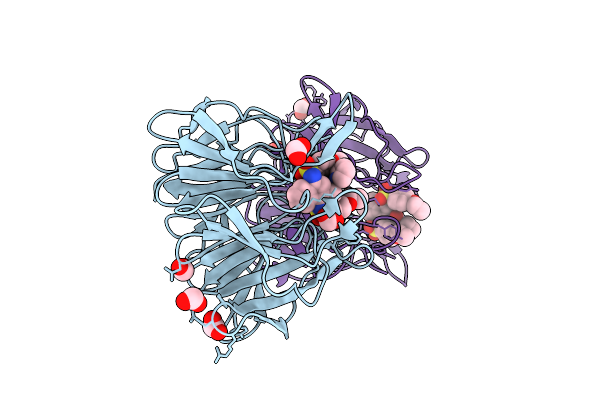 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: TRANSCRIPTION Ligands: A1L9D, FMT |
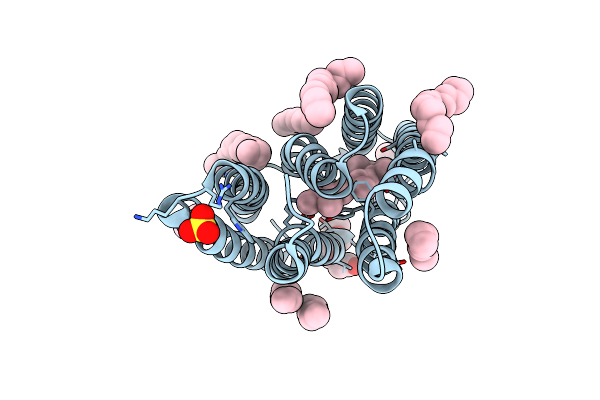 |
The Crystal Structure Of Cyanorhodopsin-Ii (Cyr-Ii) P7104R From Nodosilinea Nodulosa Pcc 7104
Organism: Nodosilinea nodulosa pcc 7104
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2023-10-25 Classification: MEMBRANE PROTEIN Ligands: RET, PG4, HEX, OCT, C14, R16, SO4, CL |
 |
Crystal Structure Of Gamma-Alpha Subunit Complex From Burkholderia Cepacia Fad Glucose Dehydrogenase In Complex With Gluconolactone
Organism: Burkholderia cepacia
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2023-09-06 Classification: OXIDOREDUCTASE Ligands: FAD, F3S, LGC |
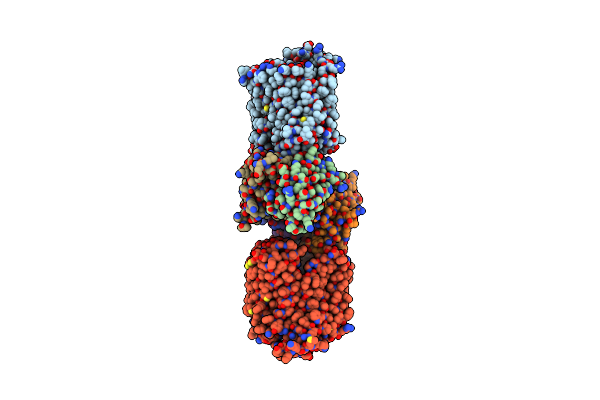 |
Crystal Structure Of The Bacterial Oxalate Transporter Oxlt In A Ligand-Free Outward-Facing Form
Organism: Oxalobacter formigenes, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2023-02-15 Classification: TRANSPORT PROTEIN/IMMUNE SYSTEM |
 |
Crystal Structure Of The Bacterial Oxalate Transporter Oxlt In An Oxalate-Bound Occluded Form
Organism: Oxalobacter formigenes, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-02-15 Classification: TRANSPORT PROTEIN/IMMUNE SYSTEM Ligands: OXL |
 |
Complex Structure Of Catalytic, Small, And A Partial Electron Transfer Subunits From Burkholderia Cepacia Fad Glucose Dehydrogenase
Organism: Burkholderia cepacia
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-12-14 Classification: OXIDOREDUCTASE Ligands: FAD, F3S, HEM |
 |
Organism: Phanerochaete chrysosporium
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2022-02-09 Classification: HYDROLASE Ligands: NAG, GOL |
 |
Organism: Phanerochaete chrysosporium
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2022-02-09 Classification: HYDROLASE Ligands: 1PE, PEG, EDO, NAG, XYP |
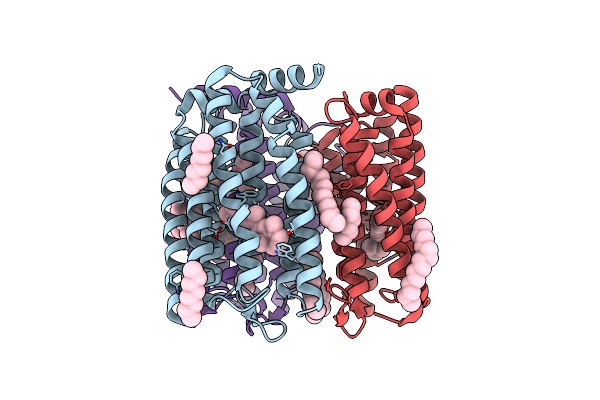 |
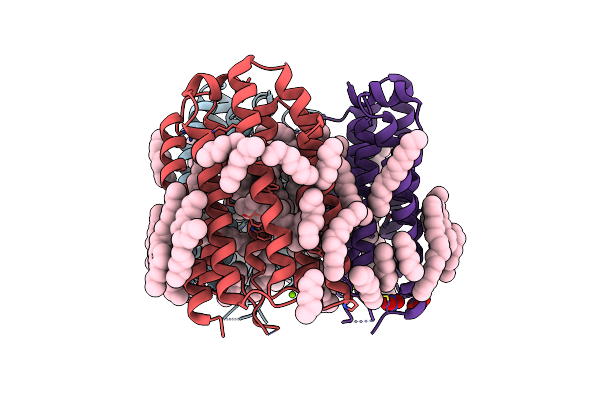
The Crystal Structure Of Cyanorhodopsin (Cyr) N2098R From Cyanobacteria Calothrix Sp. Nies-2098
Organism: Calothrix sp. nies-2098
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2020-10-21 Classification: MEMBRANE PROTEIN Ligands: RET, HEX, OCT, C14, D10 |
 |
The Crystal Structure Of Cyanorhodopsin (Cyr) N4075R From Cyanobacteria Tolypothrix Sp. Nies-4075
Organism: Tolypothrix sp. nies-4075
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-10-21 Classification: MEMBRANE PROTEIN Ligands: RET, MG, HEX, OCT, D10, D12, R16, C14, NO3 |
 |
Organism: Bacillus sp. tar1
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-09-02 Classification: HYDROLASE Ligands: MPD, CA, NI |
 |
Organism: Bacillus sp. tar1
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2020-09-02 Classification: HYDROLASE Ligands: MPD, CA, NI |
 |
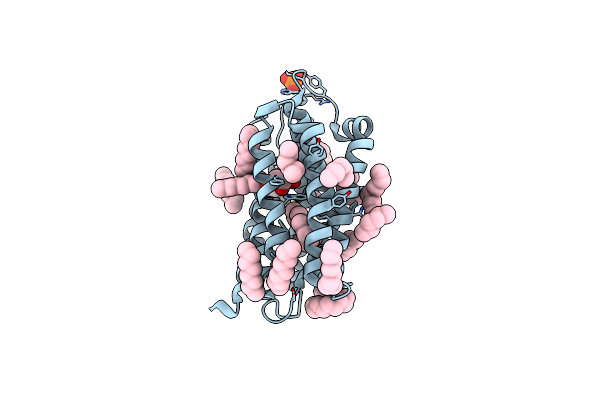
Organism: Rubrobacter xylanophilus (strain dsm 9941 / nbrc 16129)
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2020-04-08 Classification: TRANSPORT PROTEIN Ligands: MPG, RET, SO4 |
 |
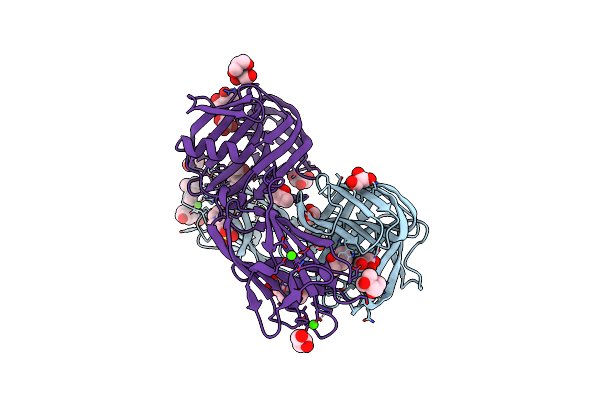
Crystal Structure Of The Dts-Motif Rhodopsin From Phaeocystis Globosa Virus 12T
Organism: Phaeocystis globosa virus 12t
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-10-02 Classification: MEMBRANE PROTEIN Ligands: RET, OCT, PO4, HEX, D10, D12, DD9, OLB |
 |
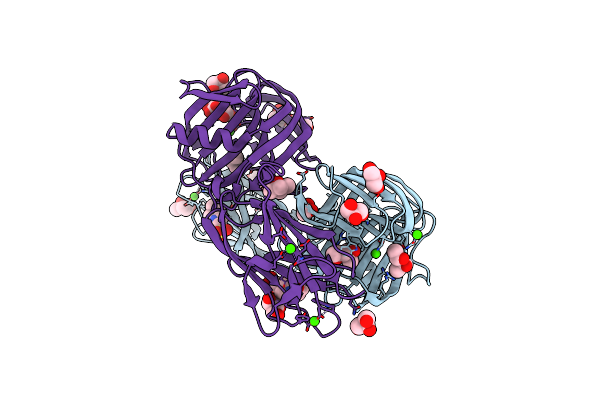
Organism: Bacillus sp. 41m-1
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2019-09-04 Classification: HYDROLASE Ligands: CA, EPE, MPD, GOL |
 |
Organism: Bacillus sp. 41m-1
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2019-09-04 Classification: HYDROLASE Ligands: CA, EPE, GOL, MPD, CIT |
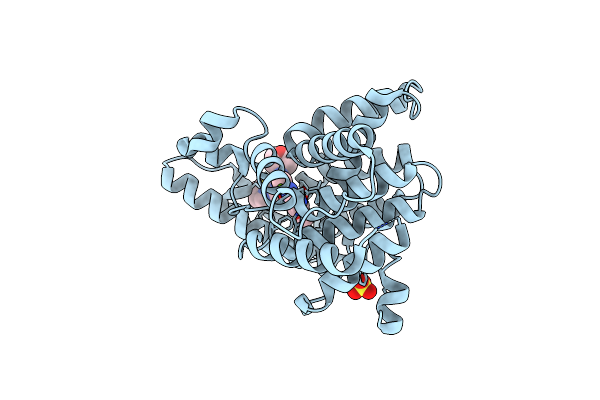 |
Discovery Of Pyrazolo[1,5-A]Pyrimidine Derivative As A Highly Selective Pde10A Inhibitor
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2019-07-17 Classification: HYDROLASE Ligands: ZN, MG, D4L, SO4 |
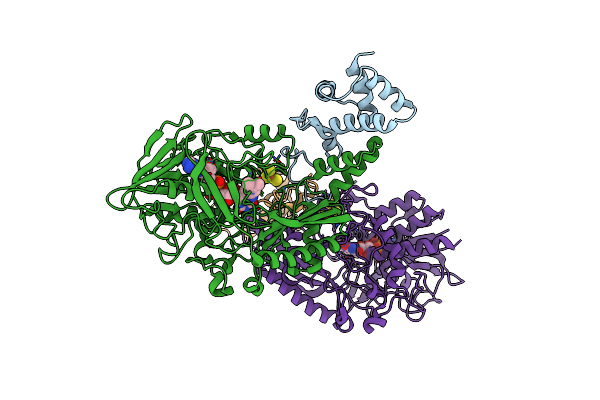 |
Crystal Structure Of Gamma-Alpha Subunit Complex From Burkholderia Cepacia Fad Glucose Dehydrogenase
Organism: Burkholderia cepacia
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-06-19 Classification: SIGNALING PROTEIN/OXIDOREDUCTASE Ligands: FAD, F3S |
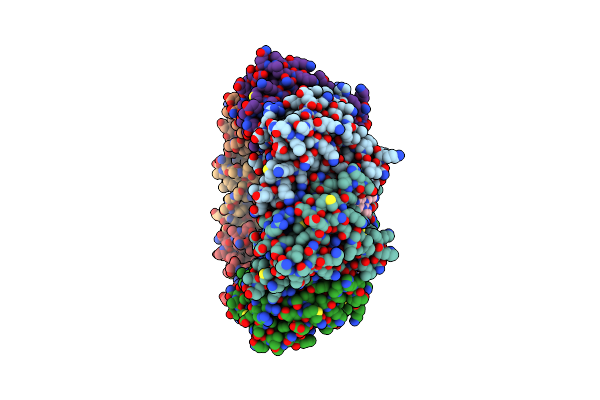 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-05-08 Classification: HYDROLASE Ligands: ONC |
 |
Organism: Aspergillus flavus nrrl3357
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2015-09-02 Classification: OXIDOREDUCTASE Ligands: FDA |

