Search Count: 101
 |
Crystal Structure Of The Actinobacillus Minor Nm305 Glucosyltransferase In Complex With Udp
Organism: Actinobacillus minor nm305
Method: X-RAY DIFFRACTION Release Date: 2025-01-15 Classification: TRANSFERASE Ligands: URI, MES |
 |
Organism: Actinobacillus minor nm305
Method: X-RAY DIFFRACTION Release Date: 2025-01-15 Classification: TRANSFERASE |
 |
Crystal Structure Of Anti-Peg Antibody M9 Fv-Clasp Fragment With Peg (Co-Crystallization With Peg3350)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2024-03-20 Classification: IMMUNE SYSTEM Ligands: 12P |
 |
Crystal Structure Of Anti-Peg Antibody M9 Fv-Clasp Fragment With Peg (Co-Crystallization With Peg2000Mme)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2024-03-20 Classification: IMMUNE SYSTEM Ligands: 12P |
 |
Crystal Structure Of Anti-Peg Antibody M9 Fv-Clasp Fragment With Peg (Co-Crystallization With Peg550Dme)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-03-20 Classification: IMMUNE SYSTEM Ligands: HZA |
 |
Crystal Structure Of Anti-Peg Antibody M11 Fv-Clasp Fragment With Peg (Co-Crystallization With Peg3350)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2024-03-20 Classification: IMMUNE SYSTEM Ligands: SO4, PE8 |
 |
Crystal Structure Of The C-Terminal Fragment (Residues 756-982 With The C864S Mutation) Of Arabidopsis Thaliana Chup1
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-01-17 Classification: PLANT PROTEIN |
 |
Crystal Structure Of The C-Terminal Fragment (Residues 716-982) Of Arabidopsis Thaliana Chup1
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-01-17 Classification: PLANT PROTEIN |
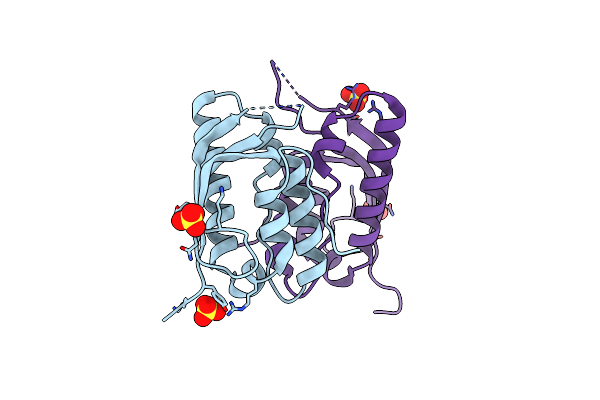 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2022-05-18 Classification: LIPID BINDING PROTEIN Ligands: SO4 |
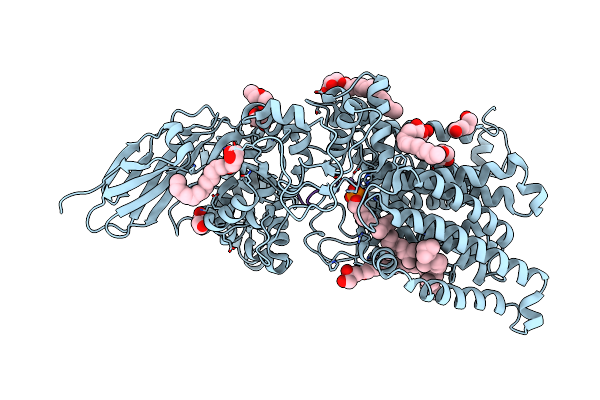 |
Archaeal Oligosaccharyltransferase Aglb From Archaeoglobus Fulgidus In Complex With An Inhibitory Peptide And A Dolichol-Phosphate
Organism: Archaeoglobus fulgidus dsm 4304, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2021-09-08 Classification: TRANSFERASE Ligands: MN, J06, PEG, 7E8 |
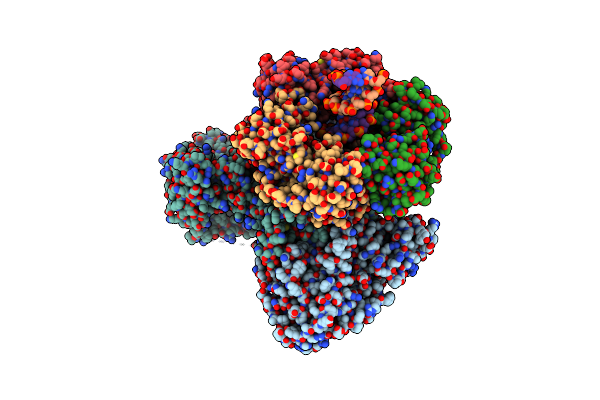 |
Organism: Thermococcus kodakarensis (strain atcc baa-918 / jcm 12380 / kod1), Thermococcus kodakarensis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-08-05 Classification: REPLICATION/DNA Ligands: FE, ZN |
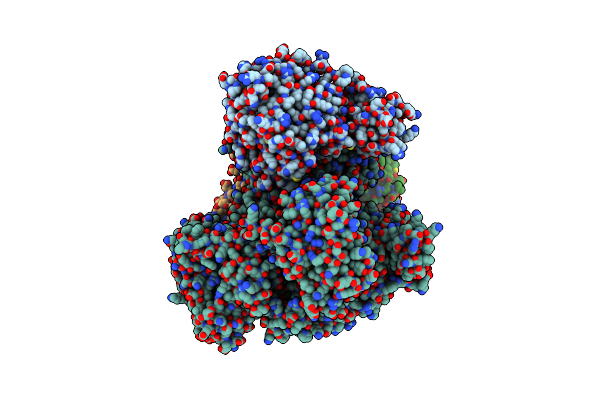 |
Organism: Thermococcus kodakarensis kod1, Thermococcus kodakarensis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-08-05 Classification: REPLICATION/DNA Ligands: FE, ZN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-10-16 Classification: LIPID BINDING PROTEIN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.76 Å Release Date: 2019-10-16 Classification: LIPID BINDING PROTEIN |
 |
Crystal Structure Of Mbpapo-Tim21 Fusion Protein With A 16-Residue Helical Linker
Organism: Escherichia coli (strain k12), Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-09-18 Classification: SUGAR BINDING PROTEIN, TRANSLOCASE |
 |
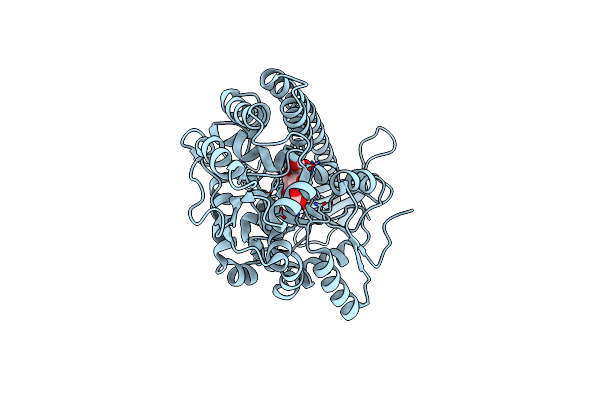
Crystal Structure Of Mbpapo-Tim21 Fusion Protein With A 17-Residue Helical Linker
Organism: Escherichia coli (strain k12), Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2019-09-18 Classification: SUGAR BINDING PROTEIN, TRANSLOCASE |
 |
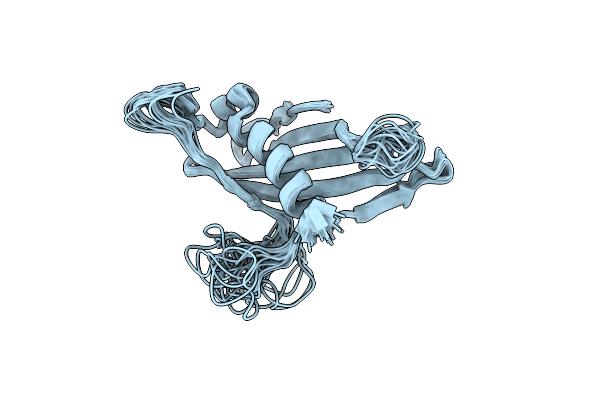
Crystal Structure Of Mbpholo-Tim21 Fusion Protein With A 17-Residue Helical Linker
Organism: Escherichia coli (strain k12), Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-09-18 Classification: SUGAR BINDING PROTEIN, TRANSLOCASE |
 |
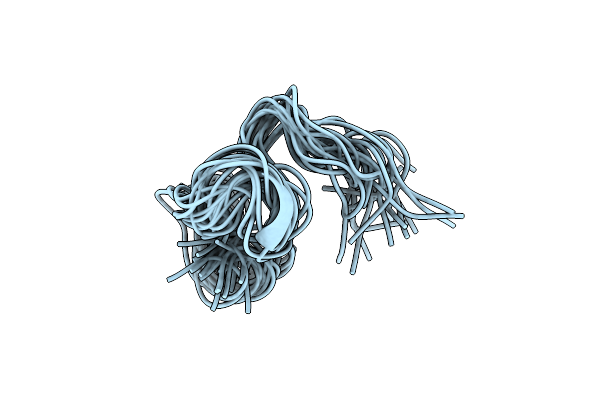
Solution Structure Of The Intermembrane Space Domain Of The Mitochondrial Import Protein Tim21 From S. Cerevisiae
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: SOLUTION NMR Release Date: 2019-09-11 Classification: PROTEIN TRANSPORT |
 |
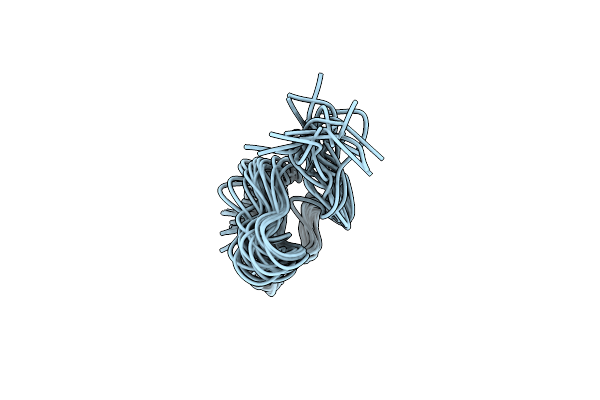
Organism: Staphylococcus warneri
Method: SOLUTION NMR Release Date: 2018-11-28 Classification: ANTIBIOTIC |
 |
Organism: Staphylococcus warneri
Method: SOLUTION NMR Release Date: 2018-11-28 Classification: ANTIBIOTIC |

