Search Count: 54
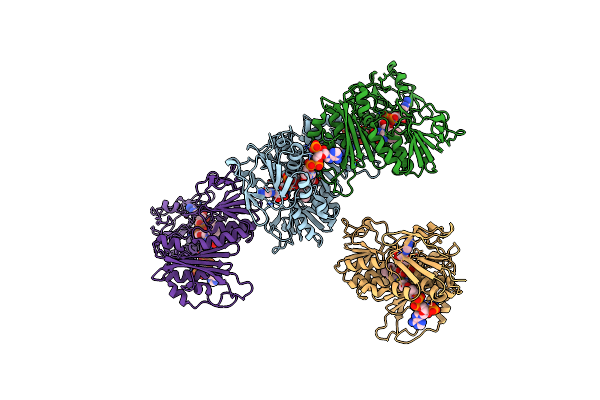 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2019-09-25 Classification: OXIDOREDUCTASE Ligands: FAD, COA |
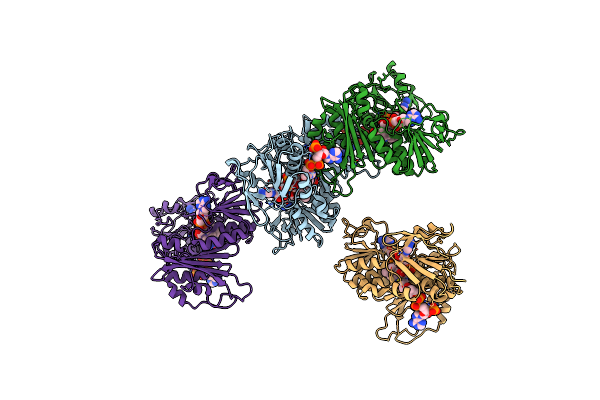 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2019-09-25 Classification: OXIDOREDUCTASE Ligands: FAD, NAD, COA |
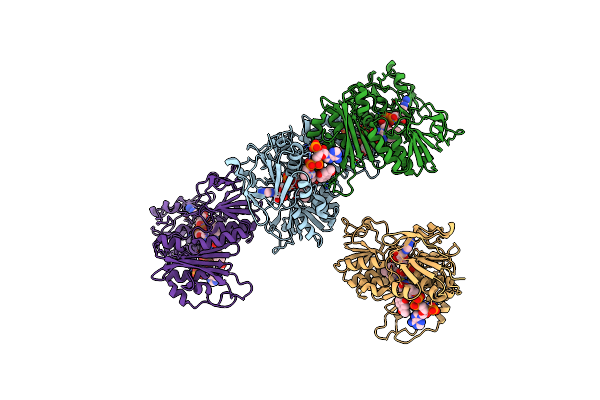 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-09-25 Classification: OXIDOREDUCTASE Ligands: FAD, COA, VK3 |
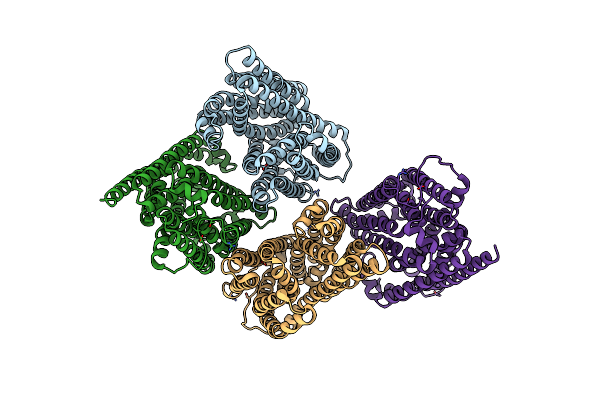 |
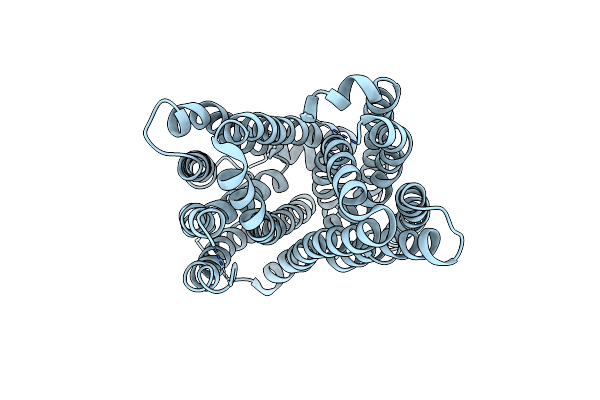
Outward-Facing Conformation Of A Multidrug Resistance Mate Family Transporter Of The Mop Superfamily.
Organism: Pyrococcus furiosus (strain atcc 43587 / dsm 3638 / jcm 8422 / vc1)
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2019-06-12 Classification: MEMBRANE PROTEIN Ligands: CS |
 |
Outward-Facing Conformation Of A Multidrug Resistance Mate Family Transporter Of The Mop Superfamily.
Organism: Pyrococcus furiosus (strain atcc 43587 / dsm 3638 / jcm 8422 / vc1)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-06-05 Classification: MEMBRANE PROTEIN |
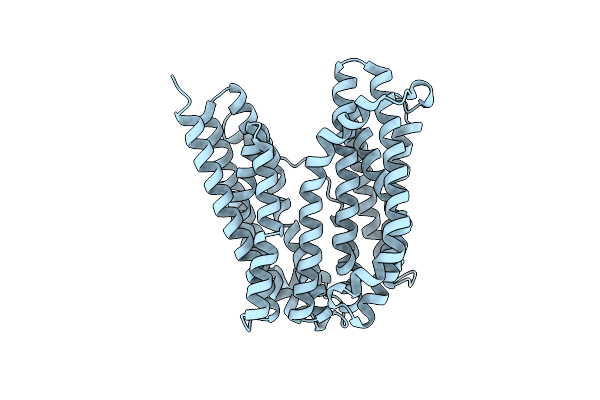 |
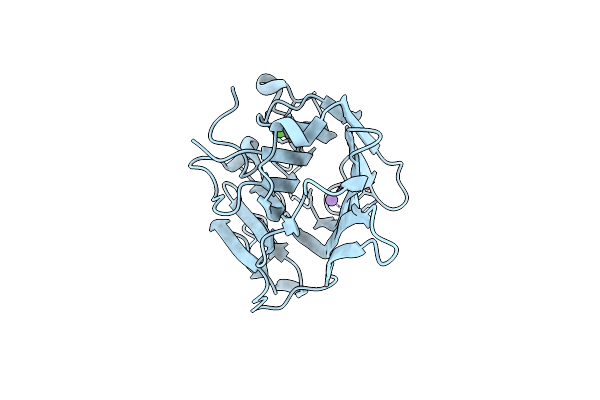
Inward-Facing Conformation Of A Multidrug Resistance Mate Family Transporter Of The Mop Superfamily.
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-05-08 Classification: MEMBRANE PROTEIN |
 |
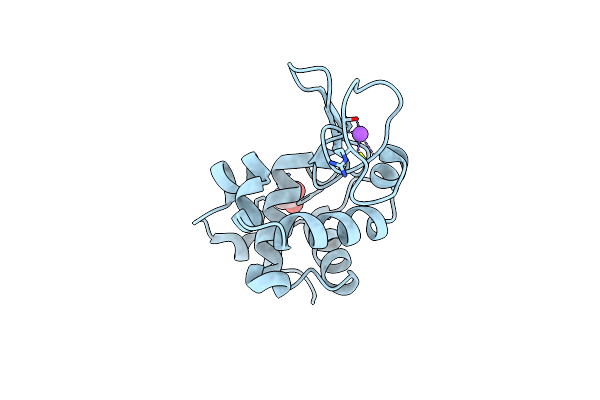
Organism: Canavalia ensiformis
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2017-12-13 Classification: SUGAR BINDING PROTEIN Ligands: CA, MN, NA, CL |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2017-12-13 Classification: HYDROLASE |
 |
Reverse Polarity Of Binding Pocket Suggests Different Function Of A Mop Superfamily Transporter From Pyrococcus Furiosus Vc1 (Dsm3638)
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2013-09-25 Classification: TRANSPORT PROTEIN Ligands: CXE, CL |
 |
Organism: Loligo vulgaris
Method: X-RAY DIFFRACTION Resolution:0.85 Å Release Date: 2011-08-17 Classification: HYDROLASE Ligands: GOL, MES, EDO, PGE, DXE, MXE, PEG, ME2, CA |
 |
High Resolution Crystal Structure Of Paracoccus Denitrificans Cytochrome C Oxidase
Organism: Paracoccus denitrificans, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2009-06-23 Classification: OXIDOREDUCTASE Ligands: HEA, CU1, MN, CA, LDA, LMT, PEO |
 |
A D-Pathway Mutation Decouples The Paracoccus Denitrificans Cytochrome C Oxidase By Altering The Side Chain Orientation Of A Distant, Conserved Glutamate
Organism: Paracoccus denitrificans, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2008-09-30 Classification: OXIDOREDUCTASE/IMMUNE SYSTEM Ligands: HEA, CU, MG, CA, LDA, LMT, PER |
 |
Structure Of Inactive Mutant Of Strictosidine Glucosidase In Complex With Strictosidine
Organism: Rauvolfia serpentina
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2008-02-05 Classification: HYDROLASE Ligands: S55 |
 |
Organism: Rauvolfia serpentina
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2008-02-05 Classification: HYDROLASE |
 |
X-Ray High Resolution Structure Of The Photosynthetic Reaction Center From Rb. Sphaeroides At Ph 8 In The Neutral State
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2007-07-03 Classification: ELECTRON TRANSPORT Ligands: GOL, BCL, LDA, BPH, U10, PO4, HTO, FE, SPO, CDL, PC1, GGD |
 |
X-Ray High Resolution Structure Of The Photosynthetic Reaction Center From Rb. Sphaeroides At Ph 8 In The Charge-Separated State
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2007-07-03 Classification: ELECTRON TRANSPORT Ligands: GOL, BCL, LDA, BPH, U10, PO4, HTO, FE, SPO, CDL |
 |
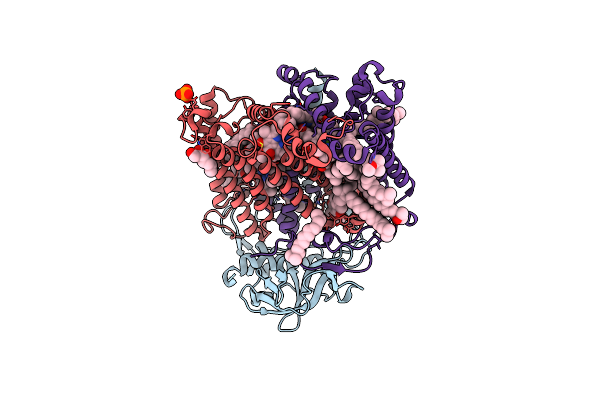
X-Ray High Resolution Structure Of The Photosynthetic Reaction Center From Rb. Sphaeroides At Ph 6.5 In The Charge-Separated State
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2007-07-03 Classification: PHOTOSYNTHESIS Ligands: GOL, LDA, BCL, BPH, UQ2, PO4, FE, U10, SPO |
 |
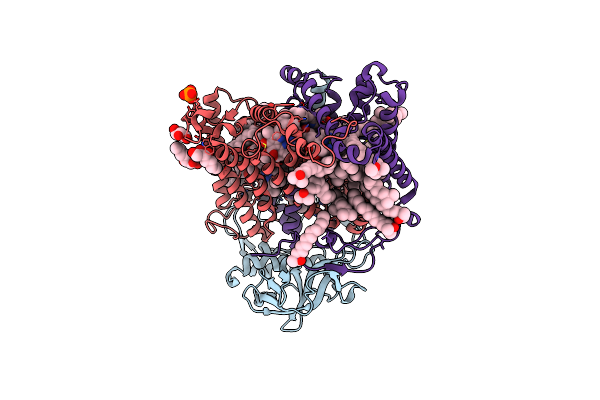
X-Ray High Resolution Structure Of The Photosynthetic Reaction Center From Rb. Sphaeroides At Ph 6.5 In The Charge-Separated State 2Nd Dataset
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-07-03 Classification: PHOTOSYNTHESIS Ligands: GOL, BCL, BPH, UQ2, PO4, HTO, LDA, FE, U10, SPO |
 |
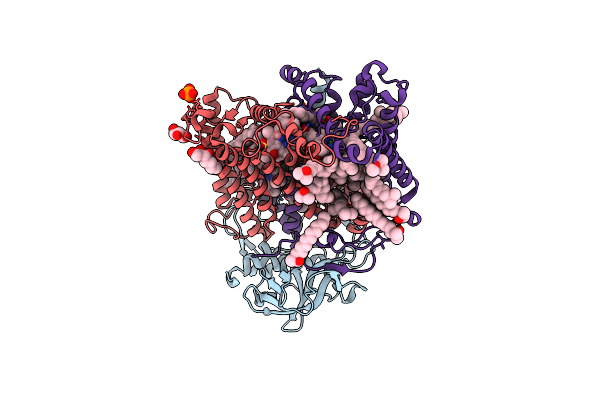
X-Ray High Resolution Structure Of The Photosynthetic Reaction Center From Rb. Sphaeroides At Ph 6.5 In The Neutral State, 2Nd Dataset
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2007-07-03 Classification: PHOTOSYNTHESIS Ligands: GOL, BCL, LDA, BPH, UQ2, PO4, HTO, FE, U10, SPO, CDL |
 |
X-Ray High Resolution Structure Of The Photosynthetic Reaction Center From Rb. Sphaeroides At Ph 6.5 In The Charge-Separated State, 3Rd Dataset
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2007-07-03 Classification: PHOTOSYNTHESIS Ligands: GOL, BCL, LDA, BPH, UQ2, PO4, HTO, FE, U10, SPO |

