Search Count: 13
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2025-03-19 Classification: TRANSFERASE Ligands: PLS, HZI |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-10-09 Classification: TRANSFERASE Ligands: PLS, A1L3N |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2024-10-09 Classification: TRANSFERASE Ligands: PGE |
 |
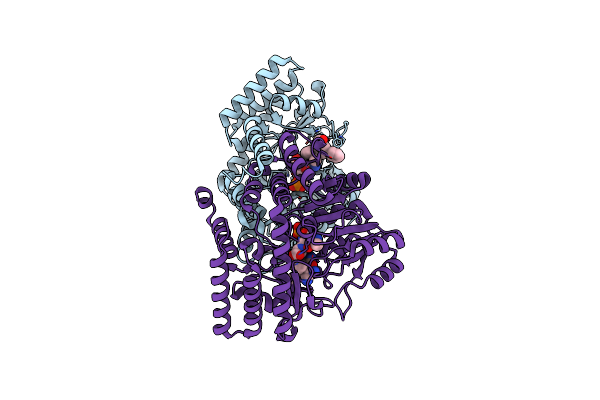
Complex Structure Of Serine Hydroxymethyltransferase From Enterococcus Faecium And Its Inhibitor
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2022-07-06 Classification: TRANSFERASE Ligands: 5M5, PLP, CL |
 |
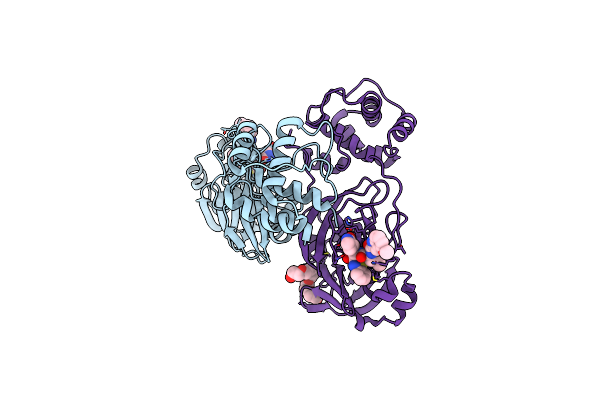
Crystal Structure Of E. Faecium Shmt In Complex With (+)-Shin-1 And Plp-Ser
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-07-06 Classification: TRANSFERASE Ligands: 5M5, PLS |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2022-07-06 Classification: TRANSFERASE Ligands: THH, PLG |
 |
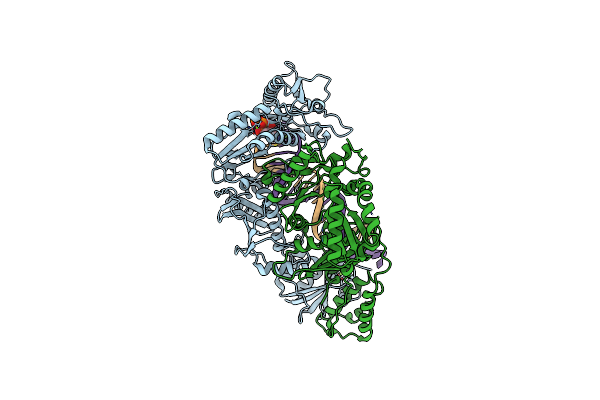
Crystal Structure Of Sars-Cov-2 Main Protease In Complex With An Inhibitor Grl-2420
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2020-09-23 Classification: VIRAL PROTEIN/INHIBITOR Ligands: V7G, 1PE |
 |
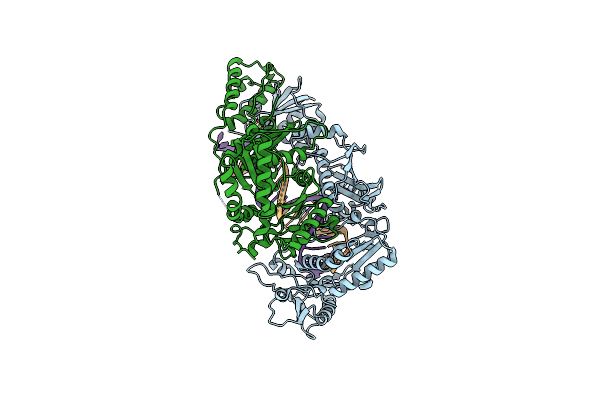
Hiv-1 Reverse Transcriptase In Complex With Dna And Efda-Triphosphate, A Translocation-Defective Rt Inhibitor
Organism: Human immunodeficiency virus type 1 group m subtype b (isolate hxb2), Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2016-08-03 Classification: transferase/dna Ligands: MG, 6FN |
 |
Hiv-1 Reverse Transcriptase In Complex With Dna That Has Incorporated Efda-Mp At The P-(Post-Translocation) Site And Dtmp At The N-(Pre-Translocation) Site
Organism: Human immunodeficiency virus type 1 group m subtype b (isolate hxb2), Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2016-08-03 Classification: transferase/dna Ligands: MG |
 |
Hiv-1 Reverse Transcriptase In Complex With Dna That Has Incorporated Efda-Mp At The P-(Post-Translocation) Site And A Second Efda-Mp At The N-(Pre-Translocation) Site
Organism: Human immunodeficiency virus type 1 group m subtype b (isolate hxb2), Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2016-08-03 Classification: transferase/dna Ligands: MG, 6FM |
 |
Hiv-1 Reverse Transcriptase In Complex With Dna That Has Incorporated A Mismatched Efda-Mp At The N-(Pre-Translocation) Site
Organism: Hiv-1 m:b_hxb2r, Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2016-08-03 Classification: transferase/dna Ligands: MG |
 |
Crystal Structure Of Hiv-1 Reverse Transcriptase (Rt) With Polymorphism Mutation K172A And K173A
Organism: Human immunodeficiency virus type 1
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2012-06-20 Classification: TRANSFERASE Ligands: MG, EDO, GOL |
 |
Crystal Structure Of The Complex Between Gp41 Fragments N36 And C34 Mutant N126K/E137Q
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2010-05-19 Classification: MEMBRANE PROTEIN Ligands: MPD, CL |

