Search Count: 19
 |
Structure Of Human Mitochondrial Rna Polymerase In Complex With Imt Inhibitor.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-30 Classification: TRANSCRIPTION Ligands: R4Q |
 |
Crystal Structure Of Degs Stress Sensor Protease In Complex With Activating Dnrlglvyqf Peptide
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-04-25 Classification: HYDROLASE Ligands: PEG, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2017-07-19 Classification: SIGNALING PROTEIN Ligands: MG, CL, NV1 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-07-19 Classification: PROTEIN BINDING Ligands: MG, CL, 8OB |
 |
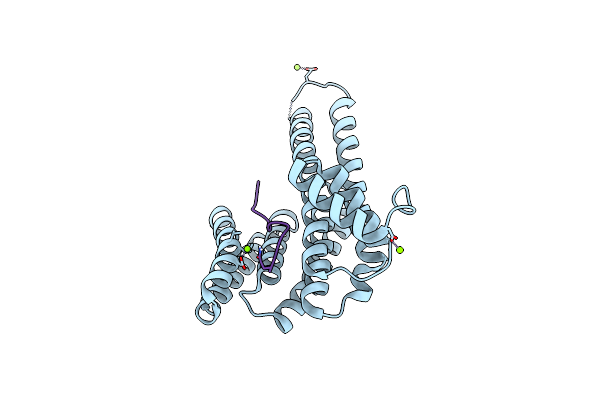
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2017-07-19 Classification: PROTEIN BINDING Ligands: MG, CL, CA, 8OE |
 |
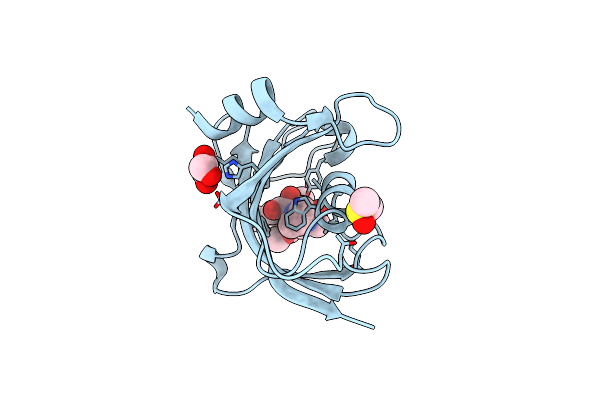
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-07-19 Classification: SIGNALING PROTEIN Ligands: MG, CL |
 |
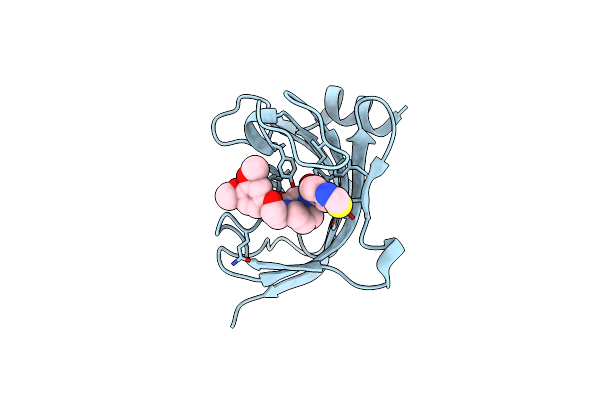
Increasing The Efficiency Efficiency Of Ligands For The Fk506-Binding Protein 51 By Conformational Control: Complex Of Fkbp51 With Compound 1-[(9S,13R,13Ar)-1,3-Dimethoxy-8-Oxo-5,8,9,10,11,12,13,13A-Octahydro-6H-9,13-Epiminoazocino[2,1-A]Isoquinolin-14-Yl]-2-(3,4,5-Trimethoxyphenyl)Ethane-1,2-Dione
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2013-08-28 Classification: ISOMERASE Ligands: 1KT, DMS, GOL |
 |
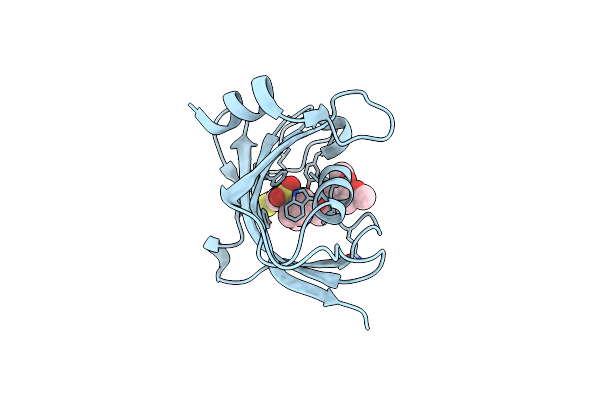
Increasing The Efficiency Efficiency Of Ligands For The Fk506-Binding Protein 51 By Conformational Control: Complex Of Fkbp51 With Compound (1S,6R)-10-(1,3-Benzothiazol-6-Ylsulfonyl)-3-[2-(3,4-Dimethoxyphenoxy)Ethyl]-3,10-Diazabicyclo[4.3.1]Decan-2-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2013-08-28 Classification: ISOMERASE Ligands: 1KU |
 |
Increasing The Efficiency Efficiency Of Ligands For The Fk506-Binding Protein 51 By Conformational Control: Complex Of Fkbp51 With (1S,6R)-3-[2-(3,4-Dimethoxyphenoxy)Ethyl]-10-[(2-Oxo-2,3-Dihydro-1,3-Benzothiazol-6-Yl)Sulfonyl]-3,10-Diazabicyclo[4.3.1]Decan-2-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2013-08-28 Classification: ISOMERASE Ligands: JFK |
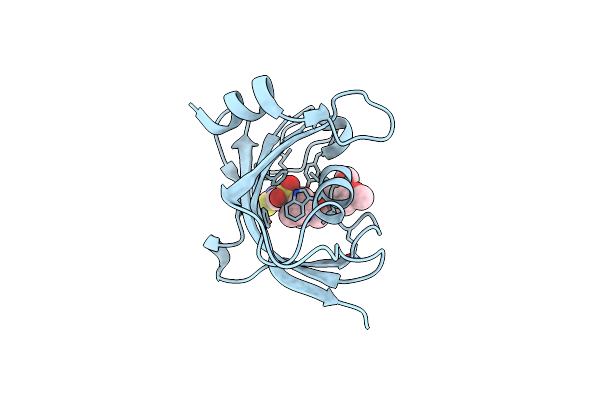 |
Increasing The Efficiency Efficiency Of Ligands For The Fk506-Binding Protein 51 By Conformational Control: Complex Of Fkbp51 With 6-({(1S,5R)-3-[2-(3,4-Dimethoxyphenoxy)Ethyl]-2-Oxo-3,9-Diazabicyclo[3.3.1]Non-9-Yl}Sulfonyl)-1,3-Benzothiazol-2(3H)-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2013-08-28 Classification: ISOMERASE Ligands: 1KY |
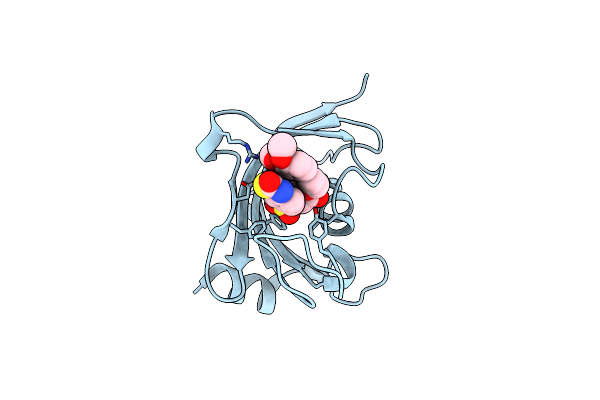 |
Increasing The Efficiency Efficiency Of Ligands For The Fk506-Binding Protein 51 By Conformational Control: Complex Of Fkbp51 With 2-(3,4-Dimethoxyphenoxy)Ethyl (2S)-1-[(2-Oxo-2,3-Dihydro-1,3-Benzothiazol-6-Yl)Sulfonyl]Piperidine-2-Carboxylate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2013-08-28 Classification: ISOMERASE Ligands: 1KZ |
 |
Structure Of Mk-3281, A Potent Non-Nucleoside Finger-Loop Inhibitor, In Complex With The Hepatitis C Virus Ns5B Polymerase
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2010-12-22 Classification: TRANSFERASE Ligands: IB8, MN |
 |
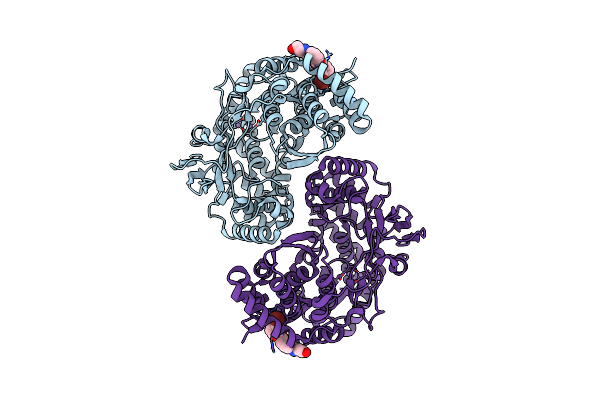
Crystal Structure Of Hepatitis C Virus Ns5B Polymerase In Complex With Thienopyrrole-Based Finger-Loop Inhibitors
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-08-18 Classification: HYDROLASE Ligands: MN, VGC |
 |
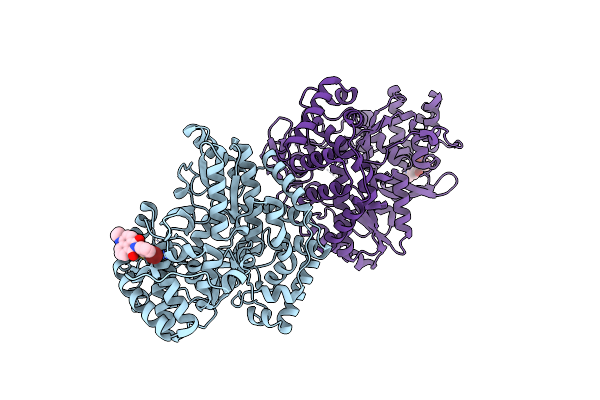
Crystal Structure Of Hepatitis C Virus Ns5B Polymerase From 1B Genotype In Complex With A Non-Nucleoside Inhibitor
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-08-11 Classification: TRANSFERASE Ligands: MN, VGI |
 |
Organism: Hepatitis c virus isolate hc-j8
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-08-04 Classification: TRANSFERASE Ligands: VGI |
 |
Nmr Structure Of Hepatitis C Virus Ns3 Serine Protease Complexed With The Non-Covalently Bound Phenethylamide Inhibitor
Organism: Hepatitis c virus
Method: SOLUTION NMR Release Date: 2009-02-03 Classification: VIRAL PROTEIN Ligands: ZN |
 |
Crystal Structure Of Hepatitis C Virus Polymerase In Complex With An Allosteric Inhibitor (Compound 1)
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2005-06-14 Classification: TRANSFERASE Ligands: MN, CMF |
 |
Crystal Structure Of Hepatitis C Virus Polymerase In Complex With An Allosteric Inhibitor (Compound 2)
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2005-06-14 Classification: TRANSFERASE Ligands: MN, POO |
 |
Crystal Structure Of The Hepatitis C Virus Ns3 Protease In Complex With A Peptidomimetic Inhibitor
Organism: Hepatitis c virus (isolate 1)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-12-09 Classification: HYDROLASE Ligands: DN1, DN2 |

