Search Count: 13
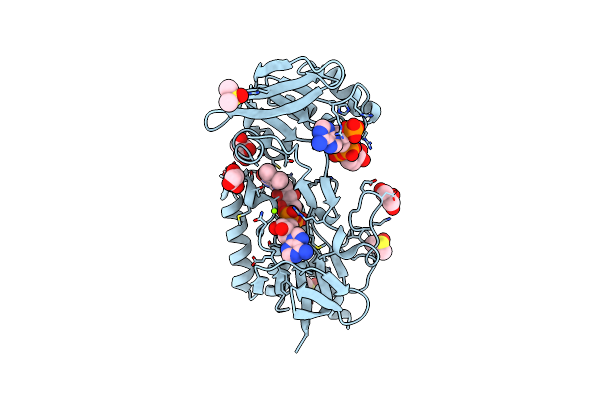 |
Crystal Structure Of Mycobacterium Smegmatis Thioredoxin Reductase In Complex With Nadph
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-02-07 Classification: OXIDOREDUCTASE Ligands: FAD, NAP, GOL, DMS, MG |
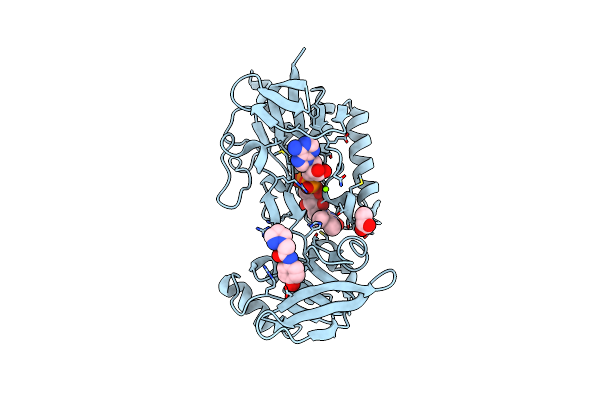 |
Crystal Structure Of Mycobacterium Smegmatis Thioredoxin Reductase In Complex With Fragment F2X-Entry H07
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2024-02-07 Classification: OXIDOREDUCTASE Ligands: FAD, GOL, U8X, MG |
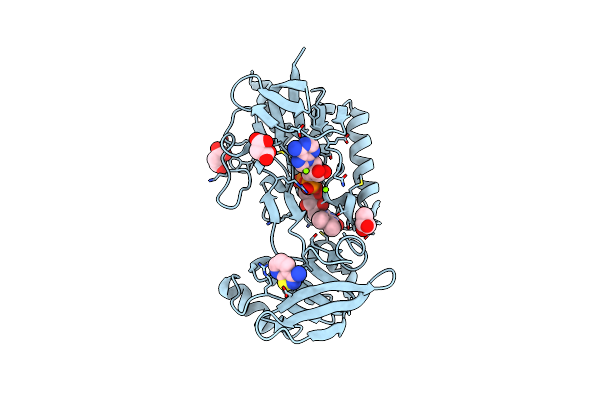 |
Crystal Structure Of Mycobacterium Smegmatis Thioredoxin Reductase In Complex With Fragment F2X-Entry A09
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-02-07 Classification: OXIDOREDUCTASE Ligands: FAD, GOL, MLI, U8O, MG |
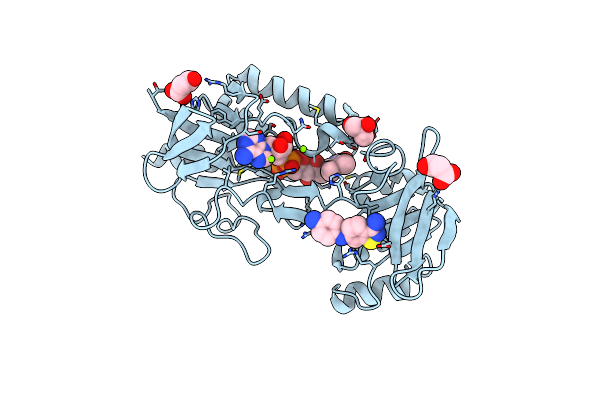 |
Crystal Structure Of Mycobacterium Smegmatis Thioredoxin Reductase In Complex With Compound 2-06
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2024-02-07 Classification: OXIDOREDUCTASE Ligands: FAD, U9C, MLI, GOL, MG, CL |
 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2023-07-26 Classification: OXIDOREDUCTASE Ligands: FAD, GOL, MLI, MG |
 |
Chlamydomonas Reinhardtii Nadph Dependent Thioredoxin Reductase 1 Domain Cs Mutant
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2022-01-19 Classification: OXIDOREDUCTASE Ligands: FAD, P4G |
 |
Crystal Structure Of Chlamydomonas Reinhardtii Nadph Dependent Thioredoxin Reductase 1 Domain
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2020-01-08 Classification: TRANSCRIPTION Ligands: EDO, LY2 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2020-01-08 Classification: TRANSCRIPTION Ligands: EGN, FMT, NA, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2020-01-08 Classification: TRANSCRIPTION Ligands: EDO, EGH |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2020-01-08 Classification: TRANSCRIPTION Ligands: EDO, FMT, DMS, SAS |
 |
Brd4 (Bd1) In Complex With Apsc-Derived Ligands (E.G. Sslh01 A Sulfasalazine Derivate)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2020-01-08 Classification: TRANSCRIPTION Ligands: EDO, DMS, EGE |
 |
Crystal Structure Of 14-3-3Zeta In Complex With A Cyclic Peptide Involving An Adamantyl And A Dicarboxy Side Chain
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2017-05-10 Classification: SIGNALING PROTEIN Ligands: BEZ |

