Search Count: 20
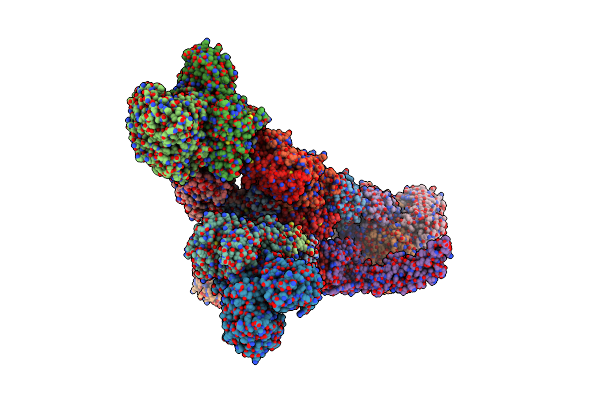 |
Formate Dehydrogenase - Heterodisulfide Reductase - Formylmethanofuran Dehydrogenase Complex From Methanospirillum Hungatei (Hexameric, Composite Structure)
Organism: Methanospirillum hungatei jf-1
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: SF4, FAD, FES, 9S8, ZN, MO, MGD |
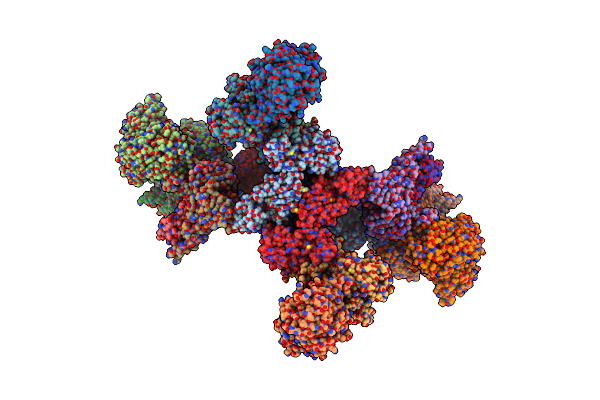 |
Formate Dehydrogenase - Heterodisulfide Reductase - Formylmethanofuran Dehydrogenase Complex From Methanospirillum Hungatei (Dimeric, Composite Structure)
Organism: Methanospirillum hungatei jf-1
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: SF4, FAD, FES, 9S8, ZN, MO, MGD |
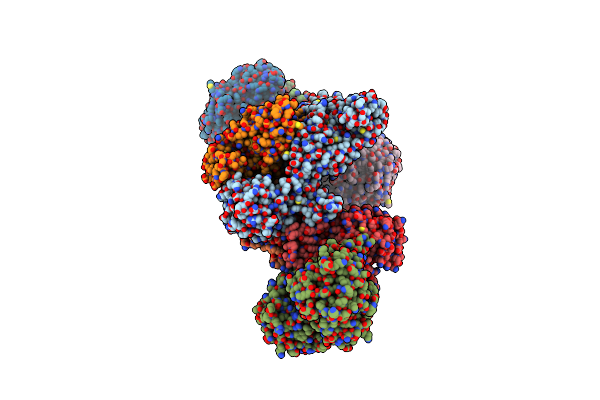 |
Formate Dehydrogenase - Heterodisulfide Reductase - Formylmethanofuran Dehydrogenase Complex From Methanospirillum Hungatei (Heterodislfide Reductase Core And Mobile Arm In Conformational State 1, Composite Structure)
Organism: Methanospirillum hungatei jf-1
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: SF4, FAD, 9S8, FES |
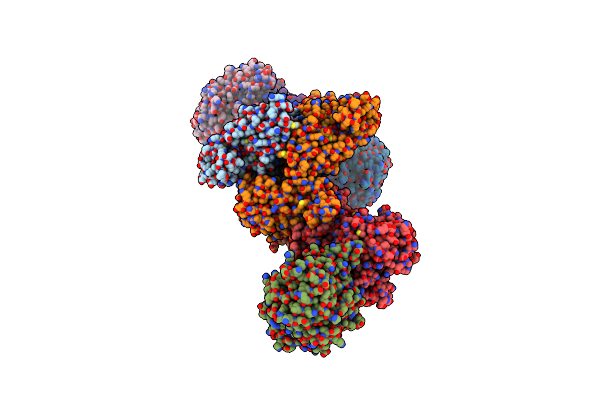 |
Formate Dehydrogenase - Heterodisulfide Reductase - Formylmethanofuran Dehydrogenase Complex From Methanospirillum Hungatei (Heterodisulfide Reductase Core And Mobile Arm In Conformational State 2, Composite Structure)
Organism: Methanospirillum hungatei jf-1
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: SF4, FAD, 9S8, FES |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2018-05-30 Classification: ANTITUMOR PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2018-05-30 Classification: ANTITUMOR PROTEIN Ligands: ZN |
 |
Heterodisulfide Reductase / [Nife]-Hydrogenase Complex From Methanothermococcus Thermolithotrophicus At 2.3 A Resolution
Organism: Methanothermococcus thermolithotrophicus dsm 2095
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-08-30 Classification: OXIDOREDUCTASE Ligands: FAD, NA, ACT, PE3, SF4, GOL, 9S8, FES, TRS, CA, MG, NFU |
 |
Heterodisulfide Reductase / [Nife]-Hydrogenase Complex From Methanothermococcus Thermolithotrophicus Soaked With Heterodisulfide For 3.5 Minutes
Organism: Methanothermococcus thermolithotrophicus dsm 2095
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-08-30 Classification: OXIDOREDUCTASE Ligands: SF4, GOL, PE3, FAD, 9S8, COM, FES, TRS, NFU, FE, ACT, TP7 |
 |
Heterodisulfide Reductase / [Nife]-Hydrogenase Complex From Methanothermococcus Thermolithotrophicus Cocrystallized With Com-Sh
Organism: Methanothermococcus thermolithotrophicus dsm 2095
Method: X-RAY DIFFRACTION Release Date: 2017-08-30 Classification: OXIDOREDUCTASE Ligands: ACT, FAD, SF4, GOL, 9S8, COM, FES, TRS, NFU, FE, PE3 |
 |
Heterodisulfide Reductase / [Nife]-Hydrogenase Complex From Methanothermococcus Thermolithotrophicus Soaked With Bromoethanesulfonate.
Organism: Methanothermococcus thermolithotrophicus dsm 2095
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-08-30 Classification: OXIDOREDUCTASE Ligands: FAD, ACT, SF4, GOL, 9S8, 9SB, FES, TRS, NFU, PE3, FE |
 |
Heterodisulfide Reductase / [Nife]-Hydrogenase Complex From Methanothermococcus Thermolithotrophicus Soaked With Heterodisulfide For 2 Minutes.
Organism: Methanothermococcus thermolithotrophicus dsm 2095
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-08-30 Classification: OXIDOREDUCTASE Ligands: FAD, SF4, GOL, 9S8, COM, SO4, FES, ACT, NFU, PE3, FE, TP7 |
 |
Organism: Methanococcus maripaludis (strain s2 / ll)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-10-26 Classification: TRANSFERASE |
 |
Hcgb From Methanocaldococcus Jannaschii With The Pyridinol Derived From Fegp Cofactor Of [Fe]-Hydrogenase
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-10-26 Classification: TRANSFERASE Ligands: 57O |
 |
Organism: Hirschia baltica (strain atcc 49814 / dsm 5838 / ifam 1418)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2016-06-22 Classification: ISOMERASE Ligands: MG |
 |
Organism: Hirschia baltica
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2016-06-22 Classification: ISOMERASE Ligands: MG, CA, CL |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2014-12-24 Classification: ISOMERASE |
 |
Organism: Rickettsia rickettsii
Method: SOLUTION NMR Release Date: 2012-05-16 Classification: RNA BINDING PROTEIN, DNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-06-10 Classification: PROTEIN BINDING Ligands: HG, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2008-06-10 Classification: PROTEIN BINDING |
 |
Crystal Structure Of The Human Fe65-Ptb1 Domain With Bound Phosphate (Trigonal Crystal Form)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2008-06-10 Classification: PROTEIN BINDING Ligands: PO4 |

