Search Count: 22
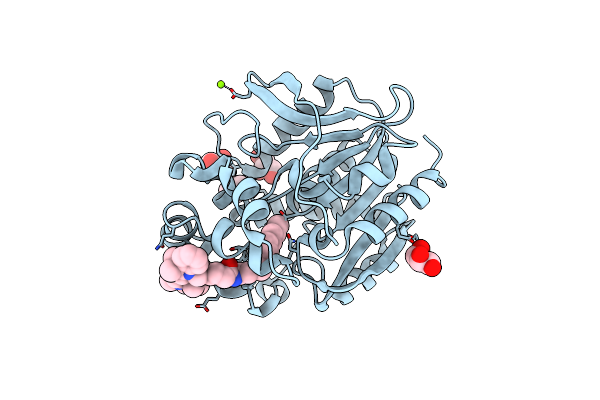 |
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7 Labeled With A Chloroalkane Cyanine3 Fluorophore Substrate
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-07-26 Classification: HYDROLASE Ligands: PJI, CL, GOL, MG |
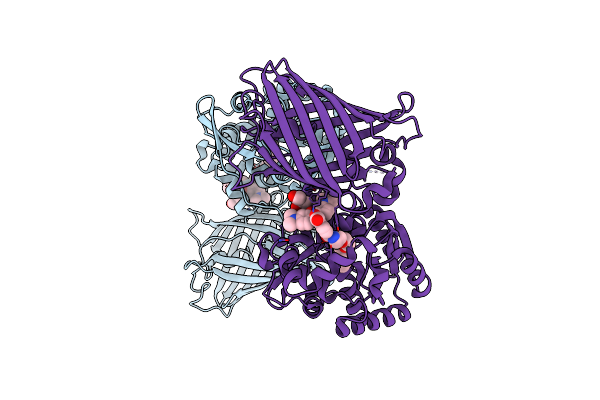 |
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7 Fusion To The Green Fluorescent Protein Gfp (Chemog1) Labeled With A Chloroalkane Tetramethylrhodamine Fluorophore Substrate
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-07-26 Classification: HYDROLASE Ligands: OEH, CL, GOL |
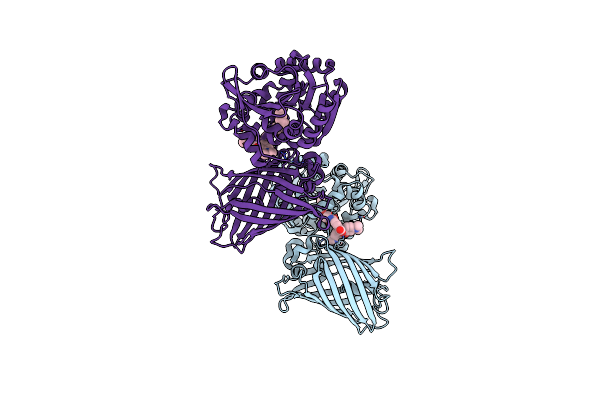 |
X-Ray Structure Of The Interface Optimized Haloalkane Dehalogenase Halotag7 Fusion To The Green Fluorescent Protein Gfp (Chemog5-Tmr) Labeled With A Chloroalkane Tetramethylrhodamine Fluorophore Substrate
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-07-26 Classification: HYDROLASE Ligands: OEH, CL |
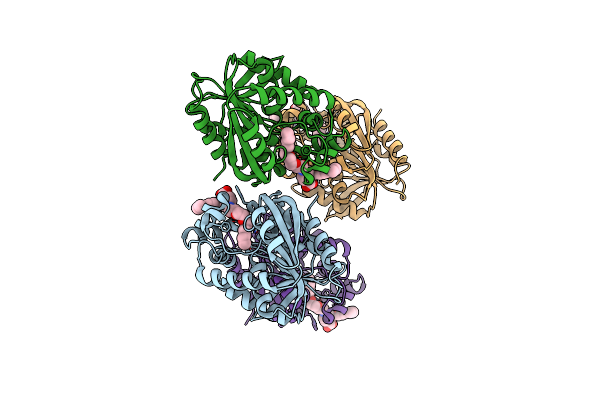 |
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7-M175W Labeled With A Chloroalkane-Tetramethylrhodamine Fluorophore Substrate
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-12-15 Classification: HYDROLASE Ligands: OEH, CL |
 |
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7-Q165W Labeled With A Chloroalkane-Tetramethylrhodamine Fluorophore Substrate
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-12-15 Classification: HYDROLASE Ligands: OEH, CL, GOL |
 |
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7-Q165H-P174R Labeled With A Chloroalkane-Tetramethylrhodamine Fluorophore Substrate
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-08-04 Classification: HYDROLASE Ligands: OEH, CL, GOL |
 |
Crystal Structure Of Gfp-Lama-F98 - A Gfp Enhancer Nanobody With Cpdhfr Insertion And Tmp And Nadph
Organism: Lama glama, Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-02-12 Classification: PROTEIN BINDING Ligands: TOP, NDP, PGE |
 |
Crystal Structure Of Gfp-Lama-G97 - A Gfp Enhancer Nanobody With Cpdhfr Insertion And Tmp And Nadph
Organism: Lama glama, Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-02-12 Classification: PROTEIN BINDING Ligands: TOP, NDP, CL, PG4, PEG |
 |
Crystal Structure Of Glutaminyl Cyclase From Drosophila Melanogaster In Space Group I4
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-09-05 Classification: HYDROLASE Ligands: ZN, EDO, SO4 |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-08-29 Classification: TRANSFERASE, HYDROLASE Ligands: ZN, PBD, GOL |
 |
Structure Of C113A/C136A Mutant Variant Of Glycosylated Glutaminyl Cyclase From Drosophila Melanogaster
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-08-29 Classification: TRANSFERASE, HYDROLASE Ligands: ZN, PBD, PEG |
 |
Crystal Structure Of Mitochondrial Isoform Of Glutaminyl Cyclase From Drosophila Melanogaster
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2012-08-29 Classification: TRANSFERASE, HYDROLASE Ligands: ZN, PBD, CL |
 |
Crystal Structure Of The C136A/C164A Variant Of Mitochondrial Isoform Of Glutaminyl Cyclase From Drosophila Melanogaster
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2012-08-29 Classification: TRANSFERASE, HYDROLASE Ligands: PBD, CL, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-06-29 Classification: TRANSFERASE Ligands: ZN, IMD, CL |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2011-06-29 Classification: TRANSFERASE Ligands: ZN |
 |
Structure Of Glycosylated Murine Glutaminyl Cyclase In Presence Of The Inhibitor Pq50 (Pdbd150)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-06-29 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: ZN, PBD, SO4, ACT |
 |
Crystal Structure Of The Enterococcus Faecalis Gluconate Specific Eiia Phosphotransferase System Component
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-09-15 Classification: TRANSFERASE Ligands: CA |
 |
Structure Of The Full-Lenght Enzyme I Of The Pts System From Staphylococcus Carnosus
Organism: Staphylococcus carnosus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2006-09-19 Classification: TRANSFERASE Ligands: SO4 |
 |
Solution Structure Of The Active-Centre Mutant Ile14Ala Of The Histidine-Containing Phosphocarrier Protein (Hpr) From Staphylococcus Carnosus
Organism: Staphylococcus carnosus
Method: SOLUTION NMR Release Date: 2005-03-08 Classification: TRANSPORT PROTEIN |
 |
Organism: Lactococcus lactis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2003-06-17 Classification: TRANSFERASE |

