Search Count: 23
 |
Crystal Structure Of The Sugar Acid Binding Protein Dctpam From Advenella Mimigardefordensis Strain Dpn7T In Complex With Xylonic Acid
Organism: Advenella mimigardefordensis dpn7
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-04-14 Classification: TRANSPORT PROTEIN Ligands: TK8 |
 |
Crystal Structure Of The Sugar Acid Binding Protein Dctpam From Advenella Mimigardefordensis Strain Dpn7T In Complex With D-Foconate
Organism: Advenella mimigardefordensis dpn7
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-04-07 Classification: SUGAR BINDING PROTEIN Ligands: TF8 |
 |
Crystal Structure Of The Sugar Acid Binding Protein Dctpam From Advenella Mimigardefordensis Strain Dpn7T In Complex With Gluconate
Organism: Advenella mimigardefordensis dpn7
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-04-07 Classification: SUGAR BINDING PROTEIN Ligands: GCO |
 |
Crystal Structure Of The Sugar Acid Binding Protein Dctpam From Advenella Mimigardefordensis Strain Dpn7T In Complex With Galactonate
Organism: Advenella mimigardefordensis dpn7
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-04-07 Classification: SUGAR BINDING PROTEIN Ligands: 2Q2 |
 |
Crystal Structure Of The Sugar Acid Binding Protein Dctpam From Advenella Mimigardefordensis Strain Dpn7T
Organism: Advenella mimigardefordensis dpn7
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2021-03-24 Classification: SUGAR BINDING PROTEIN Ligands: KDG |
 |
Organism: Pseudonocardia thermophila
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2020-12-30 Classification: HYDROLASE Ligands: PEG, PG4, MG |
 |
Organism: Pseudonocardia thermophila
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2020-12-30 Classification: HYDROLASE Ligands: HEE, 4MU, MG |
 |
Organism: Pseudomonas aestusnigri
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2020-02-26 Classification: HYDROLASE Ligands: ACT, NA |
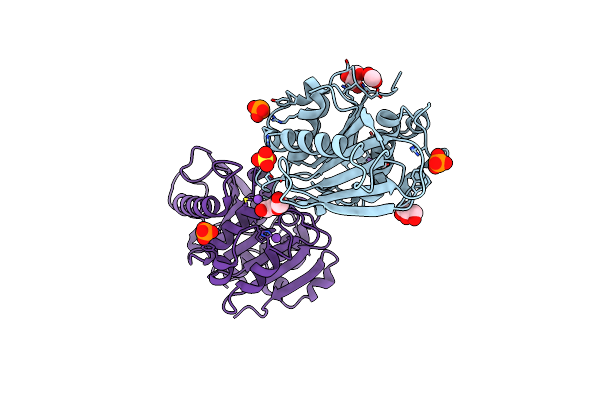 |
Organism: Pseudomonas aestusnigri
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2020-02-26 Classification: HYDROLASE Ligands: SO4, PO4, NA, CL, PEG, GOL, ACT |
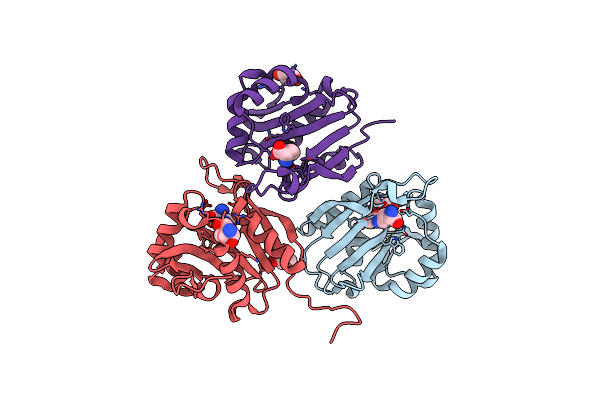 |
Diaminobutyrate Acetyltransferase Ecta From Paenibacillus Lautus In Complex With Its Product Adaba
Organism: Geobacillus sp. (strain y412mc10)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-01-29 Classification: TRANSFERASE Ligands: GOL, 9YT, TRS |
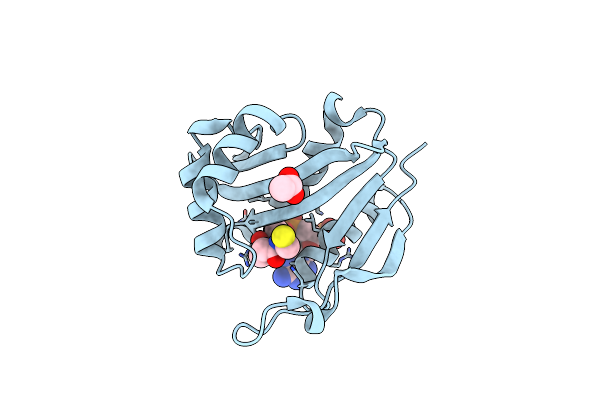 |
Diaminobutyrate Acetyltransferase Ecta From Paenibacillus Lautus In Complex With Coenzyme A
Organism: Geobacillus sp. (strain y412mc10)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-01-29 Classification: TRANSFERASE Ligands: COA, ACT |
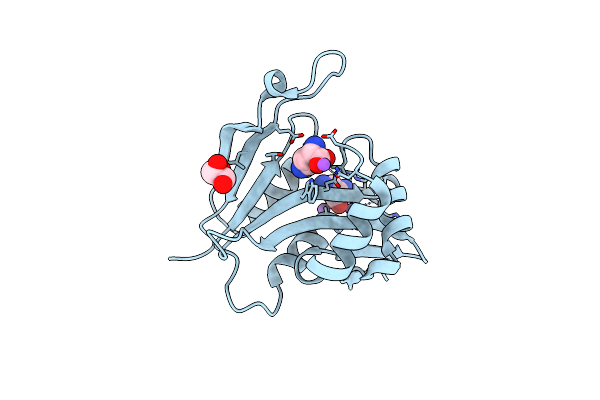 |
Diaminobutyrate Acetyltransferase Ecta From Paenibacillus Lautus In Complex With Its Substrate L-2,4-Diaminobutyric Acid (Dab)
Organism: Geobacillus sp. (strain y412mc10)
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2020-01-29 Classification: TRANSFERASE Ligands: DAB, NA, GOL |
 |
Organism: Geobacillus sp. (strain y412mc10)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-01-29 Classification: TRANSFERASE |
 |
Diaminobutyrate Acetyltransferase Ecta From Paenibacillus Lautus In Complex With Its Substrate L-2,4-Diaminobutyric Acid (Dab) And Coenzyme A
Organism: Geobacillus sp. (strain y412mc10)
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2020-01-29 Classification: TRANSFERASE Ligands: COA, DAB, MG |
 |
Structure Of Igni18, A Novel Metallo Hydrolase From The Hyperthermophilic Archaeon Ignicoccus Hospitalis Kin4/I
Organism: Ignicoccus hospitalis
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2019-10-09 Classification: HYDROLASE Ligands: ZN, PO4, K |
 |
Structural Basis For Recognition And Ring-Cleavage Of The Pseudomonas Quinolone Signal (Pqs) By Aqdc
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-07-03 Classification: HYDROLASE |
 |
Structural Basis For Recognition And Ring-Cleavage Of The Pseudomonas Quinolone Signal (Pqs) By Aqdc In Complex With Its Product
Organism: Mycobacteroides abscessus subsp. abscessus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-07-03 Classification: HYDROLASE Ligands: JWH |
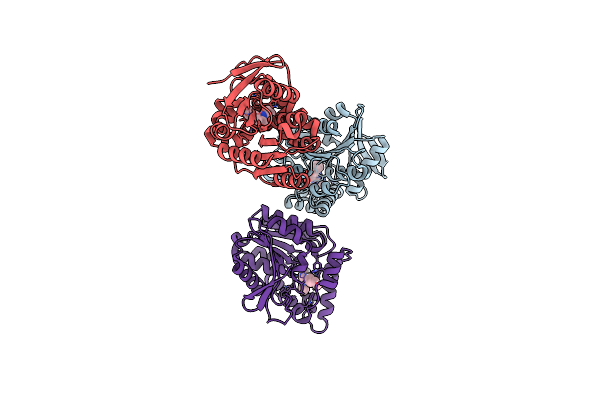 |
Structural Basis For Recognition And Ring-Cleavage Of The Pseudomonas Quinolone Signal (Pqs) By Aqdc Variant In Complex With Its Substrate
Organism: Mycobacterium abscessus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-07-03 Classification: HYDROLASE Ligands: JWW |
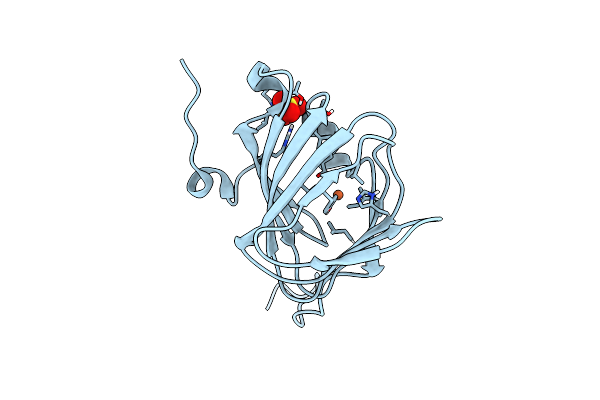 |
Organism: Paenibacillus lautus
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2018-08-22 Classification: METAL BINDING PROTEIN Ligands: FE, SO4 |
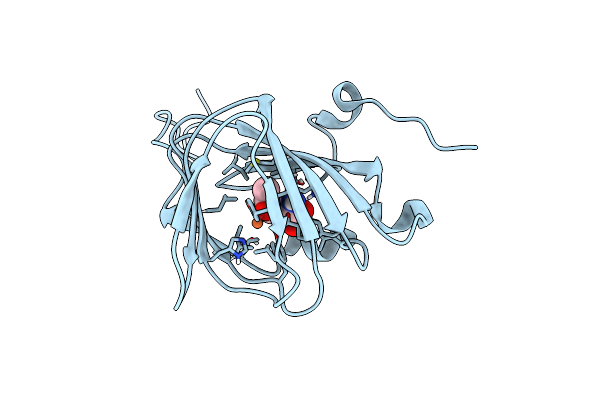 |
Organism: Paenibacillus lautus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2018-08-22 Classification: METAL BINDING PROTEIN Ligands: FE, 9YT |

