Search Count: 631
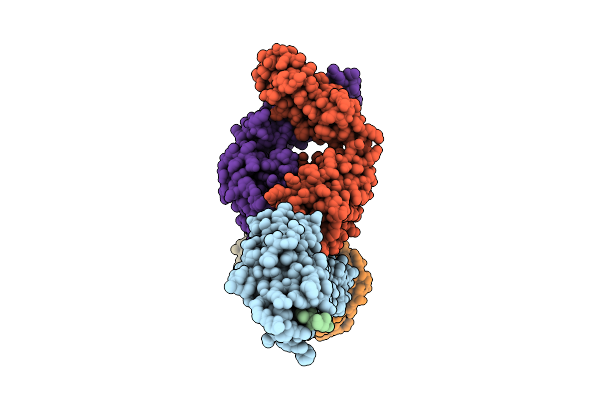 |
Antibody Fragments From Mab475 And Mab824 Bound To The Adhesin Protein Fimh
Organism: Escherichia coli, Escherichia coli k-12, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: CELL ADHESION/IMMUNE SYSTEM |
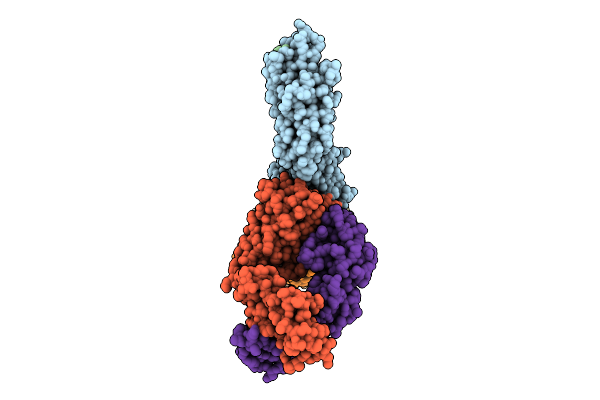 |
Antibody Fragments From Mab824 And Mab926 Bound To The Adhesin Protein Fimh
Organism: Escherichia coli, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: CELL ADHESION/IMMUNE SYSTEM |
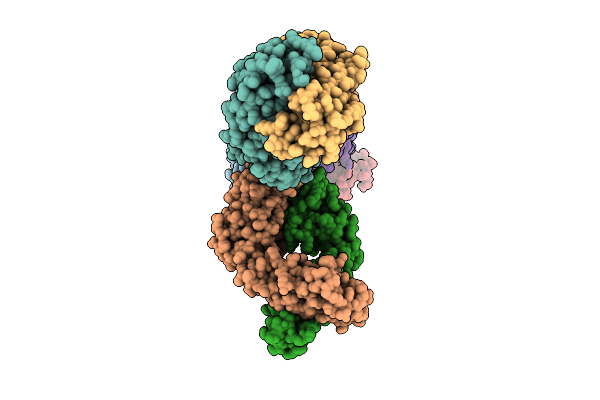 |
Antibody Fragments From Mab21, Mab475, And Mab824 Bound To The Adhesin Protein Fimh
Organism: Escherichia coli, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: CELL ADHESION/IMMUNE SYSTEM |
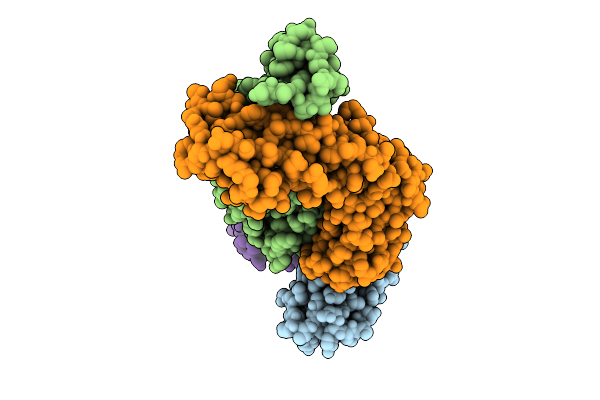 |
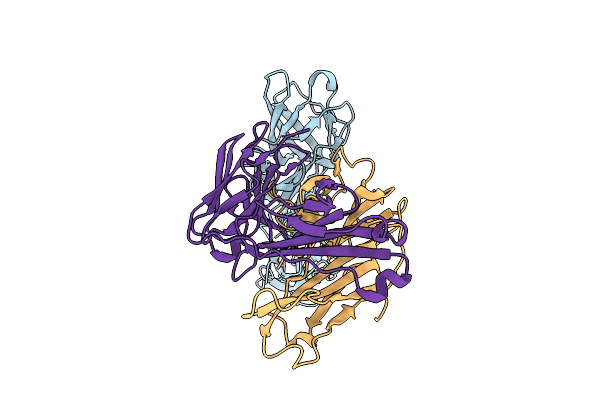
Antibody Fragments From Mab21 And Mab824 Bound To The Adhesin Protein Fimh Containing Alpha-Methyl Mannose
Organism: Escherichia coli, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: CELL ADHESION/IMMUNE SYSTEM Ligands: MMA |
 |
Organism: Escherichia coli, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: CELL ADHESION/IMMUNE SYSTEM |
 |
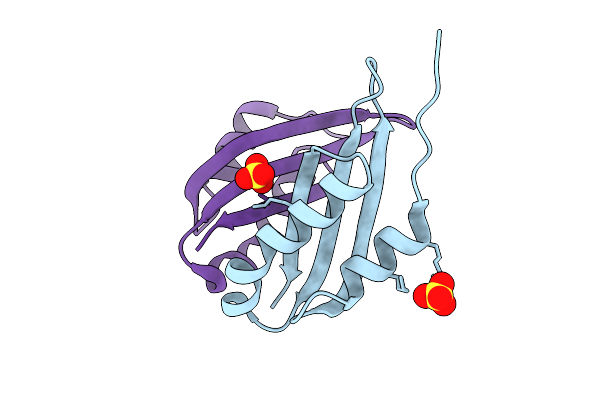
Cryo-Em Structure Of Broad Betacoronavirus Binding Antibody 1871 In Complex With Oc43 S2 Subunit
Organism: Human coronavirus oc43, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
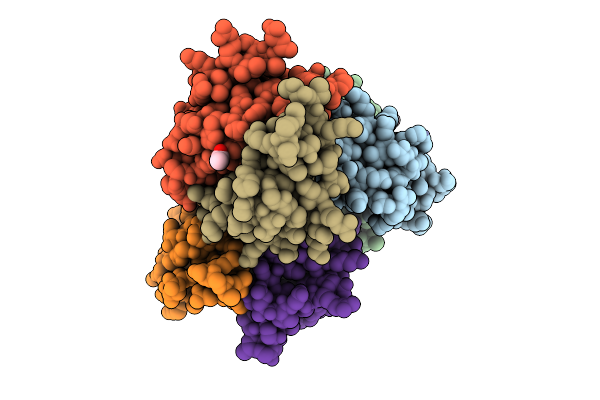
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: TRANSPORT PROTEIN Ligands: SO4 |
 |
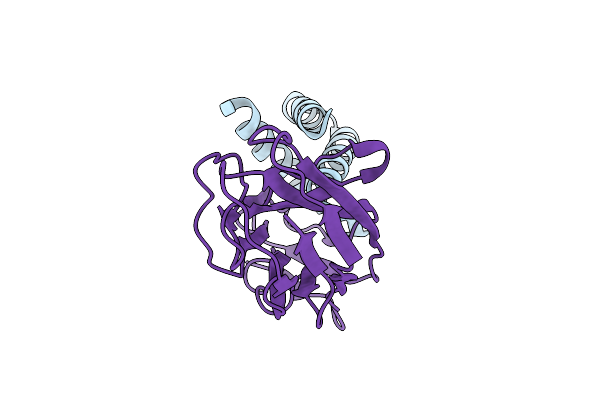
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: TRANSPORT PROTEIN Ligands: ZN, ACT |
 |
Organism: Synthetic construct, Acinetobacter baumannii acicu
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: PROTEIN BINDING |
 |
Organism: Synthetic construct, Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: DE NOVO PROTEIN |
 |
Interaction With Ak2A Links Aifm1 To Cellular Energy Metabolism. The Cryo-Em Structure Of Dimeric Aifm1 Without Any Binding Partner.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: FLAVOPROTEIN Ligands: FAD, NAD |
 |
Interaction With Ak2A Links Aifm1 To Cellular Energy Metabolism. The Cryo-Em Structure Of Dimeric Aifm1 Engaged To Mia40.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: FLAVOPROTEIN Ligands: FAD, NAD |
 |
Interaction With Ak2A Links Aifm1 To Cellular Energy Metabolism. The Cryo-Em Structure Of Dimeric Aifm1 Bound By Ak2A.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: FLAVOPROTEIN Ligands: FAD, NAD |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: TRANSFERASE Ligands: ADP, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: TRANSFERASE |
 |
Cryo-Em Structure Of E227Q Variant Of Umtck1 In Complex With Transition State Analog
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: TRANSFERASE Ligands: CRN, ADP, NO3, MG |
 |
Cryo-Em Structure Of E227Q Variant Of Umtck1 Incubated With Adp And Phosphocreatine At Ph 8.0
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: TRANSFERASE Ligands: CRN, ADP, PO4, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: TRANSFERASE Ligands: CRN, ADP, MG |
 |
Cryo-Em Structure Of Human Umtck1 In Complex With Adp And Covalent Inhibitor Cki
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: TRANSFERASE/INHIBITOR Ligands: ADP, KLU |
 |
Serine Protease Inhibitor Hciq2C1 From Heteractis Crispa With Trpa1 Mediated Antinociceptive Activity
|

