Search Count: 6
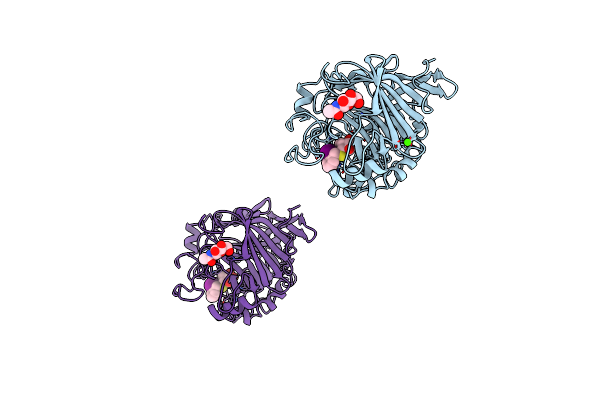 |
The Three-Dimensional Crystal Structure Of The Catalytic Core Of Cellobiohydrolase I From Trichoderma Reesei
Organism: Trichoderma reesei
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1994-11-01 Classification: HYDROLASE(O-GLYCOSYL) Ligands: NAG, BGC, CA, IBZ, GLC |
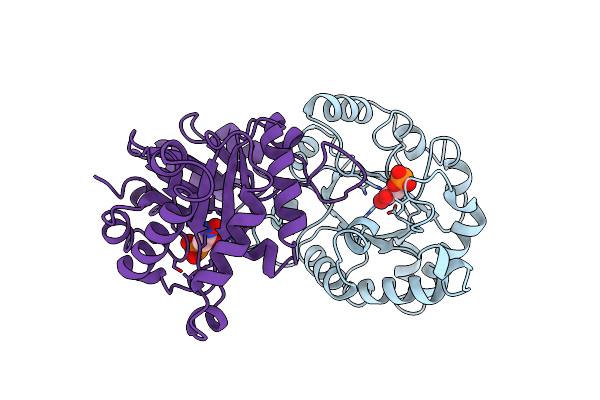 |
1.8 Angstroms Crystal Structure Of Wild Type Chicken Triosephosphate Isomerase-Phosphoglycolohydroxamate Complex
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1994-04-30 Classification: TRIOSEPHOSPHATE ISOMERASE Ligands: PGH |
 |
Electrophilic Catalysis In Triosephosphase Isomerase: The Role Of Histidine-95
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 1993-04-15 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: PGH |
 |
Organism: Hypocrea jecorina
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1991-01-15 Classification: HYDROLASE (O-GLYCOSYL) |
 |
Determination Of The Three-Dimensional Structure Of The C-Terminal Domain Of Cellobiohydrolase I From Trichoderma Reesei. A Study Using Nuclear Magnetic Resonance And Hybrid Distance Geometry-Dynamical Simulated Annealing
Organism: Hypocrea jecorina
Method: SOLUTION NMR Release Date: 1990-01-15 Classification: HYDROLASE (O-GLYCOSYL) |
 |
Determination Of The Three-Dimensional Structure Of The C-Terminal Domain Of Cellobiohydrolase I From Trichoderma Reesei. A Study Using Nuclear Magnetic Resonance And Hybrid Distance Geometry-Dynamical Simulated Annealing
Organism: Hypocrea jecorina
Method: SOLUTION NMR Release Date: 1990-01-15 Classification: HYDROLASE (O-GLYCOSYL) |

