Search Count: 16
 |
Crystal Structure Of Tailspike Depolymerase (Apk14_Gp49) From Acinetobacter Phage Vb_Abap_Apk14
Organism: Acinetobacter phage vb_abap_apk14
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-06-07 Classification: VIRAL PROTEIN Ligands: PEG, GOL, CL |
 |
Crystal Structure Of A Tailspike Depolymerase (Apk16_Gp47) From Acinetobacter Phage Apk16
Organism: Acinetobacter phage apk16
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-05-31 Classification: VIRAL PROTEIN Ligands: GOL |
 |
Crystal Structure Of Tailspike Depolymerase (Apk09_Gp48) From Acinetobacter Phage Apk09
Organism: Acinetobacter phage apk09
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2023-05-31 Classification: VIRAL PROTEIN Ligands: PEG |
 |
Crystal Structure Of Bacteriophage Cba120 Tailspike Protein 3 (Tsp3, Orf212)
Organism: Escherichia phage cba120
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2018-10-24 Classification: viral protein, hydrolase Ligands: CL |
 |
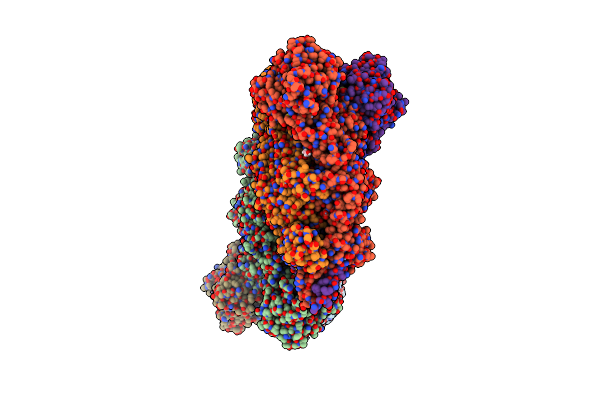
Crystal Structure Of Bacteriophage Cba120 Tailspike Protein 4 Enzymatically Active Domain (Tsp4Dn, Orf213)
Organism: Escherichia virus cba120
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2018-10-24 Classification: viral protein, hydrolase Ligands: ACT, K, NA, ZN, SO4, CL |
 |
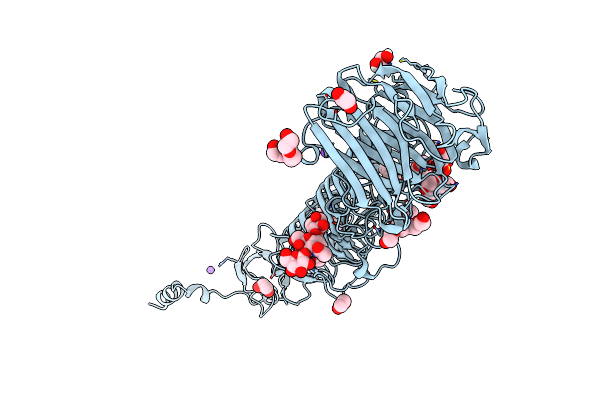
Crystal Structure Of Bacteriophage Cba120 Tailspike Protein 2 Enzymatically Active Domain (Tsp2Dn, Orf211)
Organism: Escherichia phage cba120
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2018-10-24 Classification: viral protein, hydrolase Ligands: ZN, K, EDO |
 |
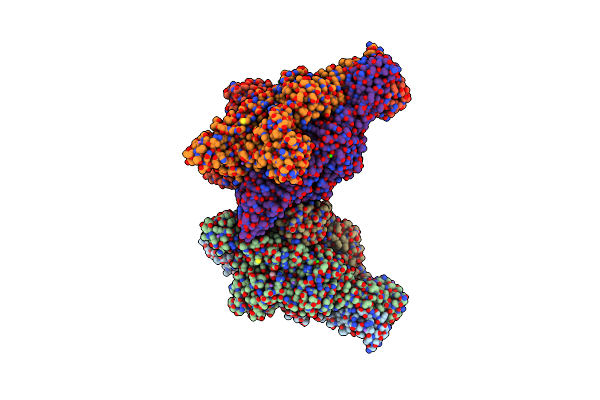
Crystal Structure Of Bacteriophage Cba120 Tailspike Protein 2 Enzymatically Active Domain (Tsp2Dn, Orf211) Complex With Escherichia Coli O157-Antigen
Organism: Escherichia phage cba120
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2018-10-24 Classification: viral protein, hydrolase Ligands: PGE, PG4, SO4, EDO, NA, K, CL |
 |
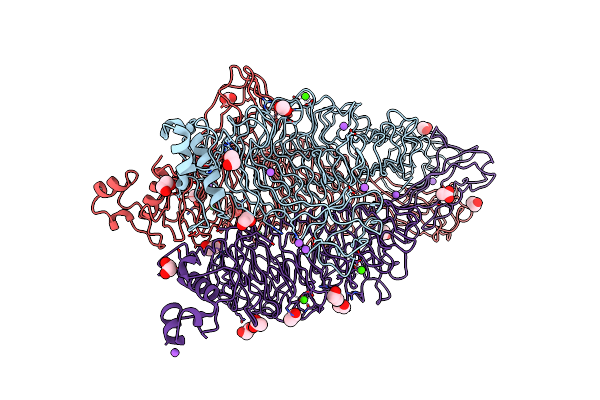
Crystal Structure Of The Tailspike Protein Gp49 From Pseudomonas Phage Lka1
Organism: Pseudomonas phage lka1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-11-18 Classification: STRUCTURAL PROTEIN Ligands: EDO, NA, CA |
 |
Organism: Pseudomonas phage phi297
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2015-11-18 Classification: LYASE Ligands: CA, NA, EDO, ACT |
 |
Organism: Escherichia phage vb_ecop_g7c
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2015-06-24 Classification: HYDROLASE Ligands: PEG, EDO, ZN, SO4, CL |
 |
Structure Of The Carbohydrate-Recognition Domain Of Human Langerin With Man5 (Man Alpha1-3(Man Alpha1-6)Man Alpha1-6)(Man- Alpha1-3)Man
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-12-08 Classification: SUGAR BINDING PROTEIN Ligands: CA, MAN |
 |
Structure Of The Carbohydrate-Recognition Domain Of Human Langerin With Man4 (Man Alpha1-3(Man Alpha1-6)Man Alpha1-6Man)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2010-12-08 Classification: SUGAR BINDING PROTEIN Ligands: MAN, CA |
 |
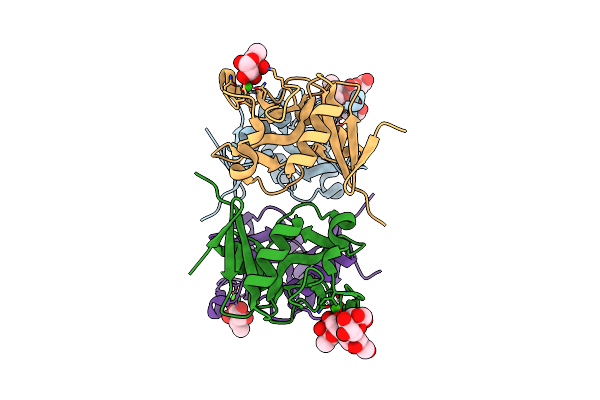
Structure Of The Carbohydrate-Recognition Domain Of Human Langerin With Man2 (Man Alpha1-2 Man)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2010-12-08 Classification: SUGAR BINDING PROTEIN Ligands: MAN, CA |
 |
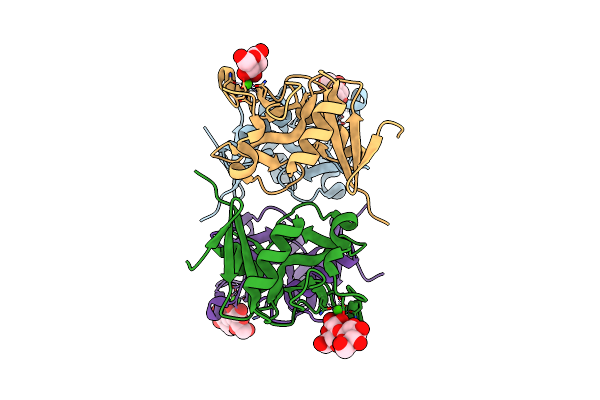
Structure Of The Carbohydrate-Recognition Domain Of Human Langerin With Blood Group B Trisaccharide (Gal Alpha1-3(Fuc Alpha1-2)Gal)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-12-08 Classification: SUGAR BINDING PROTEIN Ligands: CA, FUC |
 |
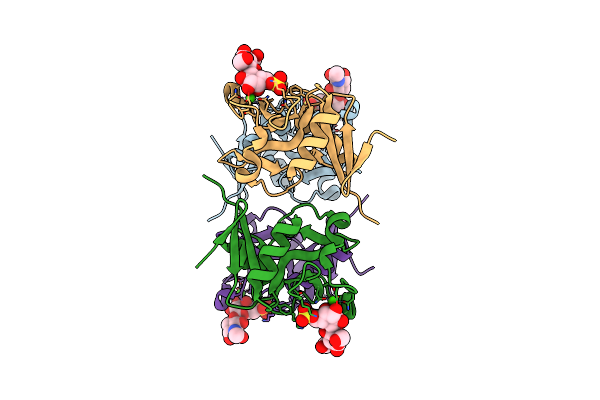
Structure Of The Carbohydrate-Recognition Domain Of Human Langerin With Laminaritriose
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2010-12-08 Classification: SUGAR BINDING PROTEIN Ligands: BGC, CA |
 |
Structure Of The Carbohydrate-Recognition Domain Of Human Langerin With 6-So4-Gal-Glcnac
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-12-08 Classification: SUGAR BINDING PROTEIN Ligands: CA |

