Search Count: 86
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: TRANSFERASE/INHIBITOR Ligands: 8X7, ZN, ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: TRANSFERASE/INHIBITOR Ligands: A1A1S, NA, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: TRANSFERASE/INHIBITOR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: TRANSFERASE/INHIBITOR |
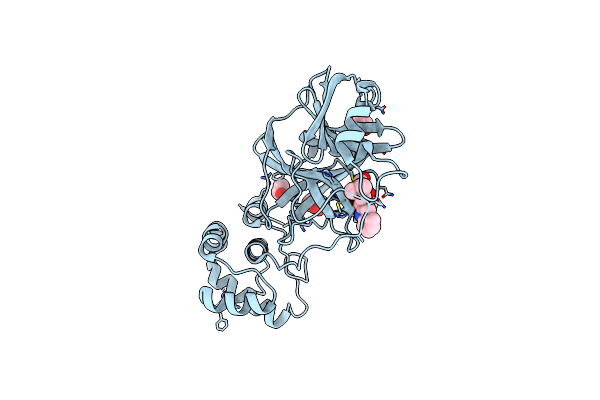 |
Structure Of Sars-Cov-2 Main Protease With Potent Peptide Aldehyde Inhibitor
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2024-08-07 Classification: HYDROLASE/INHIBITOR Ligands: A1ADM, NA, EDO, PEG |
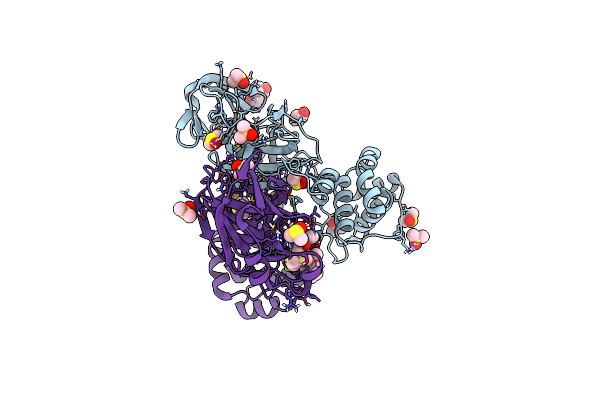 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2024-04-03 Classification: VIRAL PROTEIN Ligands: BTB, A1AD0, DMS, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-01-24 Classification: OXIDOREDUCTASE Ligands: NAI, YBO, EDO, PEG |
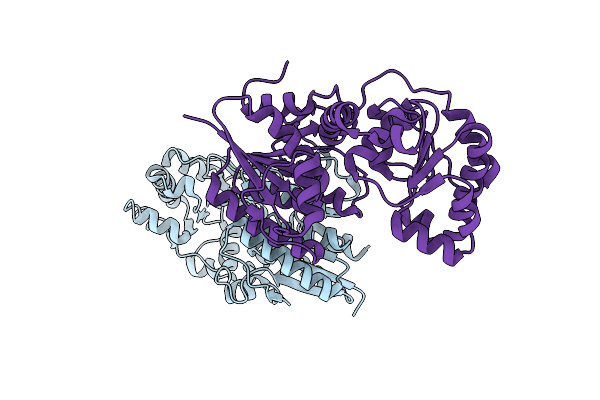 |
X-Ray Crystal Structure Of Udp- 2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid-2-Epimerase From Thermus Thermophilus Strain Hb27, Apo Form, Ph 9
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-09-13 Classification: ISOMERASE Ligands: CL |
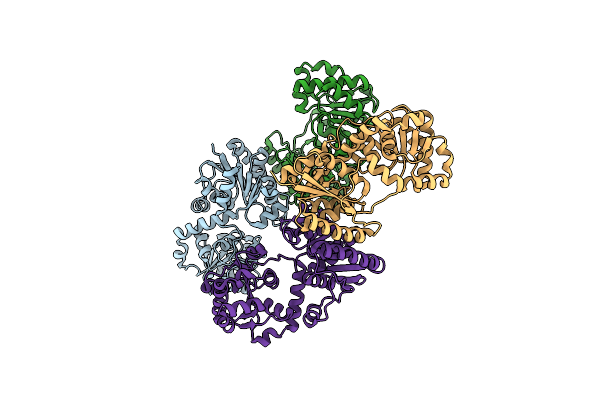 |
X-Ray Crystal Structure Of Udp- 2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid-2-Epimerase From Thermus Thermophilus Strain Hb27, D98N Mutation, Apo Structure At Ph 6
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-09-13 Classification: ISOMERASE Ligands: CL, NA |
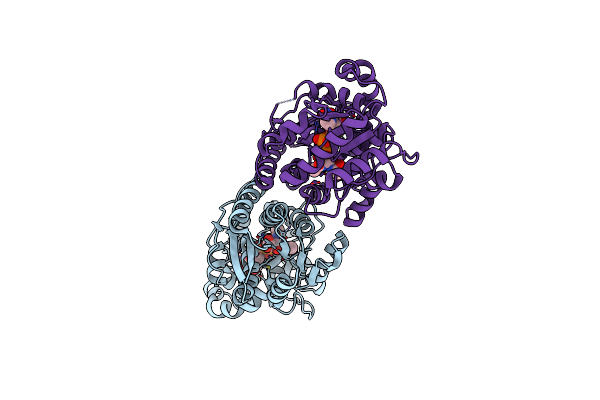 |
X-Ray Crystal Structure Of Udp- 2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid-2-Epimerase From Thermus Thermophilus Strain Hb27 In Complex With Its Product Udp-2,3-Diacetamido-2,3-Dideoxy-D-Mannuronic Acid At Ph 5
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-09-13 Classification: ISOMERASE Ligands: MJ3, CL |
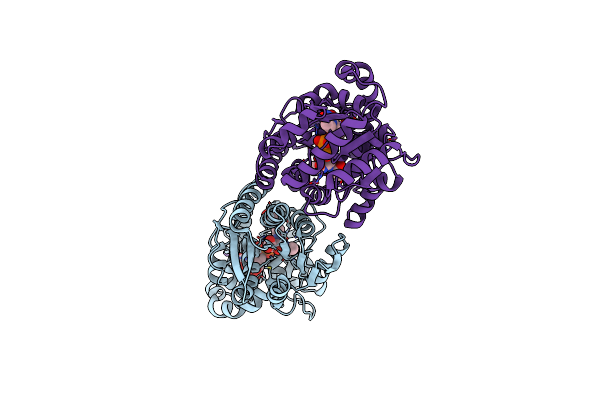 |
X-Ray Crystal Structure Of Udp- 2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid-2-Epimerase From Thermus Thermophilus Strain Hb27 In Complex With Its Product Udp-2,3-Diacetamido-2,3-Dideoxy-D-Mannuronic Acid At Ph 9
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-09-13 Classification: ISOMERASE Ligands: MJ3, EDO, NA, CL |
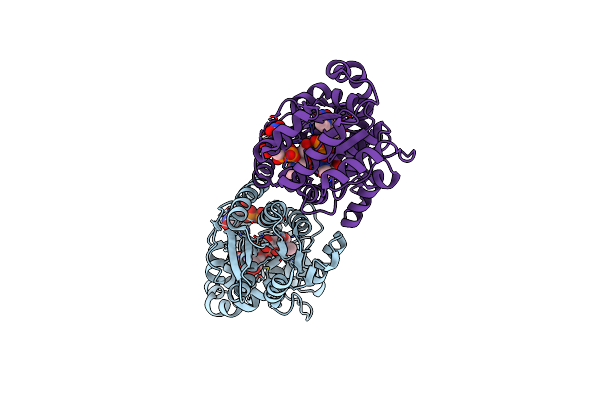 |
X-Ray Crystal Structure Of Udp-2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid-2-Epimerase From Thermus Thermophilus Strain Hb27, D98N Variant In The Presence Of Udp-2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid, Udp-N-Acetylglucosamine And Udp At Ph 7
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-09-13 Classification: ISOMERASE Ligands: MJL, UDP, UD1 |
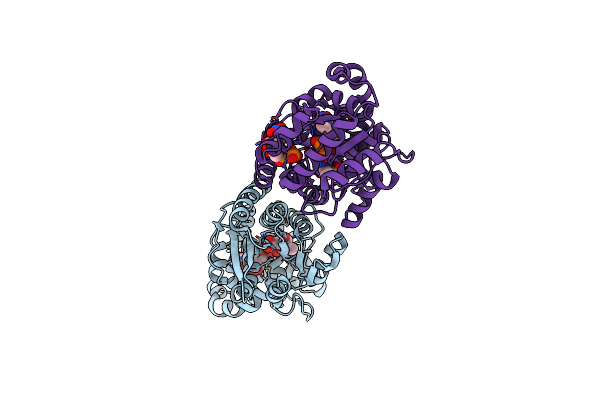 |
X-Ray Crystal Structure Of Udp-2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid-2-Epimerase From Thermus Thermophilus Strain Hb27, D98N Variant In The Presence Of Udp-2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid And Udp At Ph 9
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-09-13 Classification: ISOMERASE Ligands: MJL, UDP |
 |
X-Ray Crystal Structure Of Udp-2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid-2-Epimerase From Thermus Thermophilus Strain Hb27, D98N Variant In The Presence Of Udp-2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid And Udp-N-Acetylglucosamine At Ph 9
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-09-13 Classification: ISOMERASE Ligands: MJL, UD1, CL, NA |
 |
X-Ray Crystal Structure Of Udp-2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid-2-Epimerase From Thermus Thermophilus Strain Hb27, D98N Variant In The Presence Of Udp-2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid And Udp-N-Acetylglucosamine At Ph 6
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-09-13 Classification: ISOMERASE Ligands: MJL, UD1, CL |
 |
X-Ray Crystal Structure Of Udp-2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid-2-Epimerase From Thermus Thermophilus Strain Hb27, D98N Variant In The Presence Of Udp-2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid And Udp At Ph 6
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-09-13 Classification: ISOMERASE Ligands: MJL, UDP, CL |
 |
X-Ray Crystal Structure Of Udp-2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid-2-Epimerase From Thermus Thermophilus Strain Hb27, D98N Variant In The Presence Of Udp-2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid And Udp At Ph 8
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-09-13 Classification: ISOMERASE Ligands: MJL, UDP, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2022-11-30 Classification: CELL CYCLE Ligands: 799 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2022-11-30 Classification: CELL CYCLE Ligands: X3W |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2022-11-30 Classification: CELL CYCLE Ligands: X3R |

