Search Count: 1,068
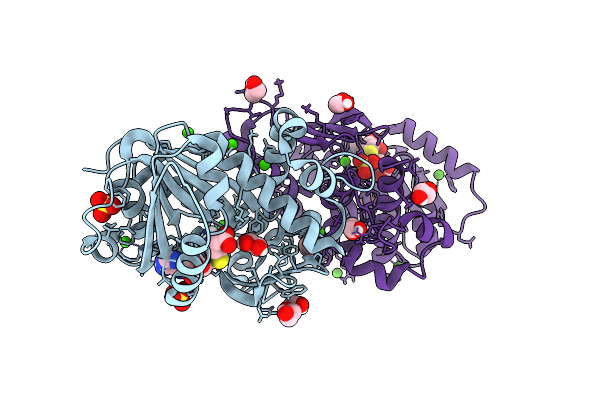 |
Structure Of Kluyveromyces Lactis Mrna Cap (Guanine-N7) Methyltransferase, Abd1, In Complex With Sah
Organism: Kluyveromyces lactis nrrl y-1140
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: SAH, EDO, PEG, SO4, CA |
 |
Structure Of Kluyveromyces Lactis Mrna Cap (Guanine-N7) Methyltransferase, Abd1, In Complex With Adenine And M7Gtp
Organism: Kluyveromyces lactis nrrl y-1140
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: ADE, MGP, P4G, SO4, EDO |
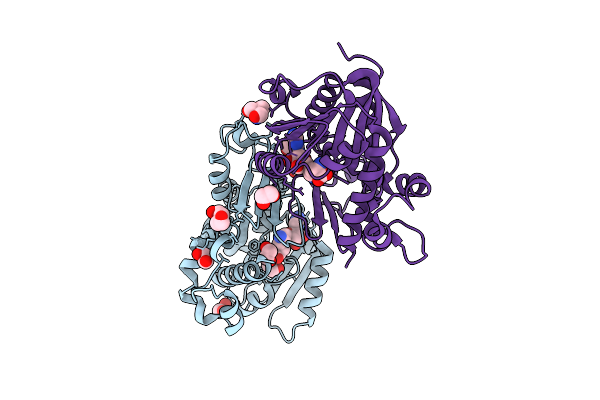 |
Structure Of Kluyveromyces Lactis Mrna Cap (Guanine-N7) Methyltransferase, Abd1, In Complex With Sinefungin
Organism: Kluyveromyces lactis nrrl y-1140
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: SFG, EDO, PEG, TRS |
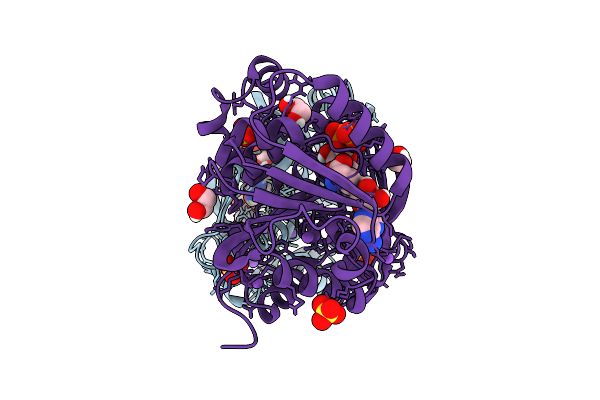 |
Structure Of Kluyveromyces Lactis Mrna Cap (Guanine-N7) Methyltransferase, Abd1, In Complex With Sinefungin And Gtp
Organism: Kluyveromyces lactis nrrl y-1140
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: SFG, GTP, NA, SO4, EDO |
 |
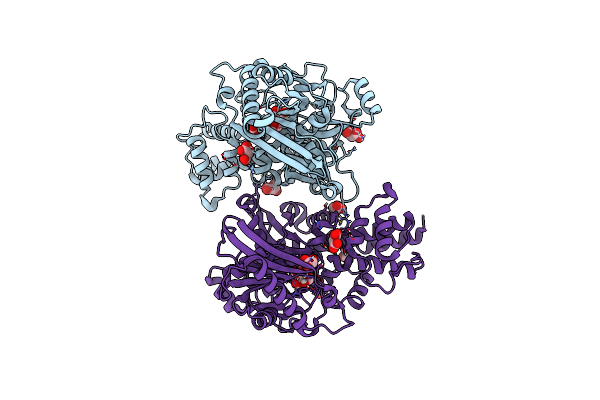
Crystal Structure Of Kluyveromyces Lactis Glucokinase In Complex With Glucose
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: BGC, MLI |
 |
Crystal Structure Of Kluyveromyces Lactis Glucokinase In Complex With Mannose
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: MAN, MLI |
 |
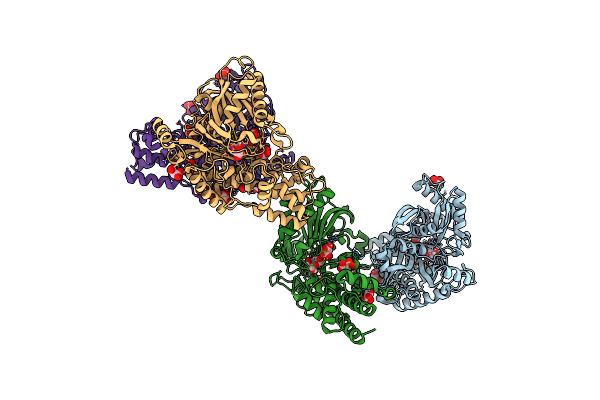
Crystal Structure Of Kluyveromyces Lactis Glucokinase Mutant H304Q In Complex With Glucose
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: BGC, MLI |
 |
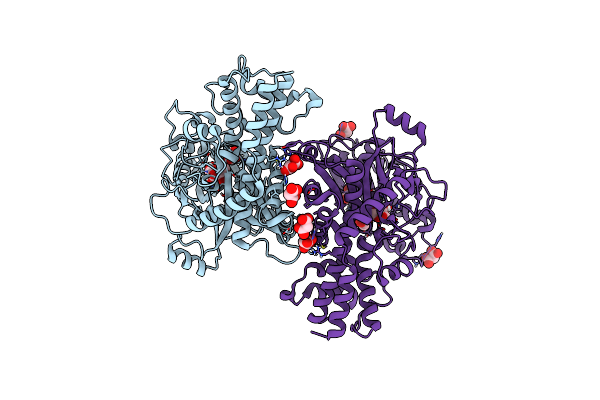
Crystal Structure Of Kluyveromyces Lactis Glucokinase Mutant H304Q In Complex With Mannose
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: MAN, MLI |
 |
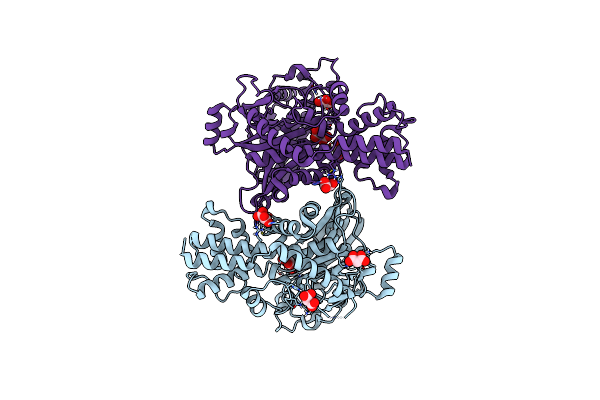
Crystal Structure Of Kluyveromyces Lactis Glucokinase Mutant H304Q In Complex With Fructose
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: FRU, MLI |
 |
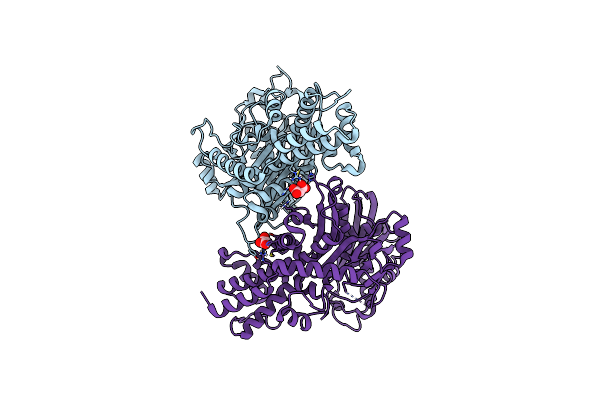
Crystal Structure Of The Intermediate Open State Of Apo Kluyveromyces Lactis Glucokinase
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: MLI |
 |
Crystal Structure Of The Open State Of Apo Kluyveromyces Lactis Glucokinase
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: MLI |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Swivelled Conformation (Model Py48S-Auc-Swiv-Eif1)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-2)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-3.1)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-3.2)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-2.1)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-2.2)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-Eif1)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Closed Conformation (Model Py48S-Auc-Eif5)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RIBOSOME Ligands: MG, ZN, MET, GCP |

