Search Count: 150
 |
Room Temperature Structure Of Kr2 Rhodopsin In Pentameric Form At 95% Relative Humidity
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN Ligands: RET, NA, OLC, LFA |
 |
Room-Temperature Structure Of Kr2 Rhodopsin In Pentameric Form At 85% Relative Humidity
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN Ligands: RET, NA, OLC, LFA |
 |
Hen Egg-White Lysozyme Structure Embedded In Lcp Medium At 95% Relative Humidity
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-07-16 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-07-16 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-07-16 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-07-16 Classification: HYDROLASE |
 |
Hen Egg-White Lysozyme Structure Collected At Euxfel Spb/Sfx With Hve Injection Method
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-07-16 Classification: HYDROLASE |
 |
Hen Egg-White Lysozyme Structure Embedded In Lcp Medium At 95% Relative Humidity
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-07-16 Classification: HYDROLASE |
 |
Hen Egg-White Lysozyme Structure Embedded In Lcp Medium At 85% Relative Humidity
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-07-16 Classification: HYDROLASE |
 |
Organism: Glycine max
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-06-04 Classification: PLANT PROTEIN Ligands: CYC |
 |
Organism: Oscillatoria acuminata
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-02-26 Classification: LYASE Ligands: FMN, GOL, CA |
 |
Organism: Glycine max
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2025-01-08 Classification: PLANT PROTEIN Ligands: CYC, 1PE, PEG, PGE |
 |
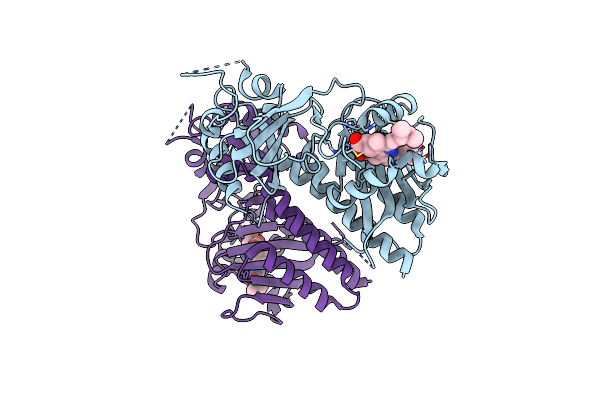
Organism: Glycine max
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2025-01-08 Classification: PLANT PROTEIN Ligands: O6E, PG4 |
 |
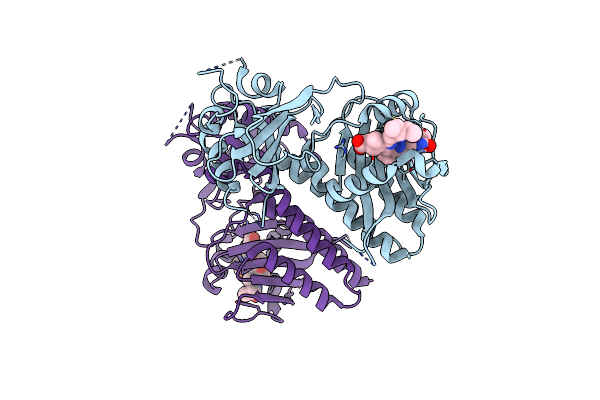
Organism: Glycine max
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-01-08 Classification: PLANT PROTEIN Ligands: CYC |
 |
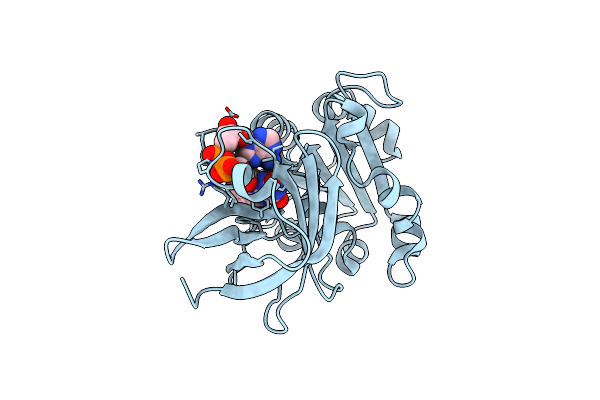
Organism: Glycine max
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-01-08 Classification: PLANT PROTEIN Ligands: CYC |
 |
Room Temperature Structure Of Fad-Containing Ferrodoxin-Nadp Reductase From Brucella Ovis At Euxfel
Organism: Brucella ovis atcc 25840
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-11-27 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
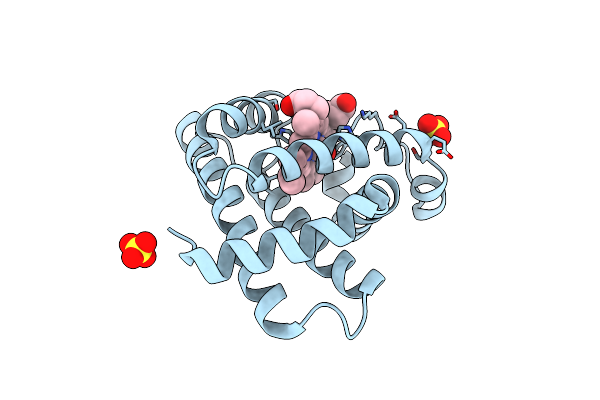
Room Temperature Structure Of Fad-Containing Ferrodoxin-Nadp Reductase From Brucella Ovis At Lcls
Organism: Brucella ovis atcc 25840
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-11-27 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Multicopy-Refined Carboxymyoglobin Photolysis Time Series At 5 Mj/Cm2 Laser Fluence, 327 Fs Time Delay
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-03-06 Classification: OXYGEN STORAGE Ligands: HEM, SO4, CMO |
 |
Carboxymyoglobin Dark State For Comparison With Power Titration And 23 / 101 Mj/Cm2 Time Series
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2024-01-31 Classification: OXYGEN STORAGE Ligands: HEM, SO4, CMO |
 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2024-01-31 Classification: OXYGEN STORAGE Ligands: HEM, SO4, CMO |

