Search Count: 121
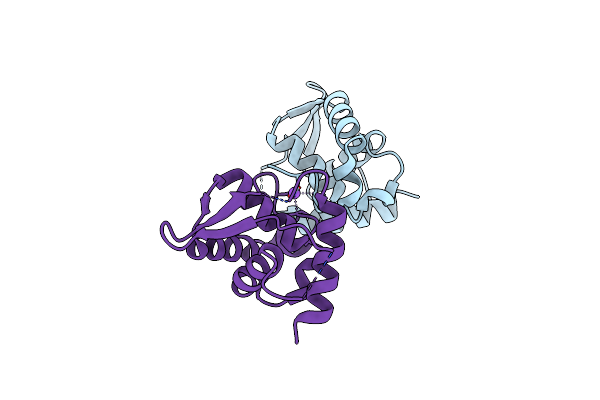 |
Crystal Structure Of A Fragment Of Nuclear Factor Related To Kappa-B-Binding Protein (Residues 370-495) (Nfrkb) From Homo Sapiens At 2.18 A Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2011-11-02 Classification: Transcription regulation, DNA BINDING |
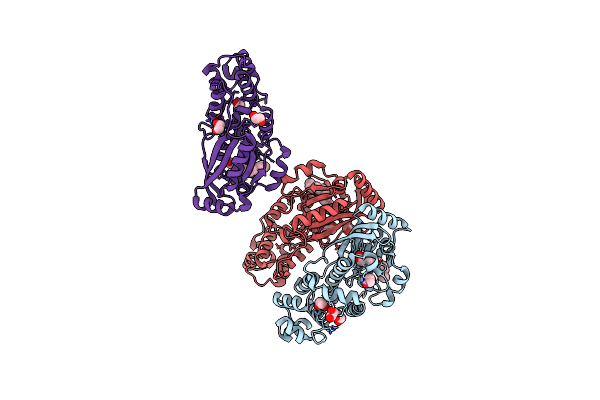 |
Crystal Structure Of A Pas And Dna Binding Domain Containing Protein (Caur_2278) From Chloroflexus Aurantiacus J-10-Fl At 2.30 A Resolution
Organism: Chloroflexus aurantiacus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-01-19 Classification: TRANSCRIPTION REGULATOR Ligands: MYR, CL, EDO |
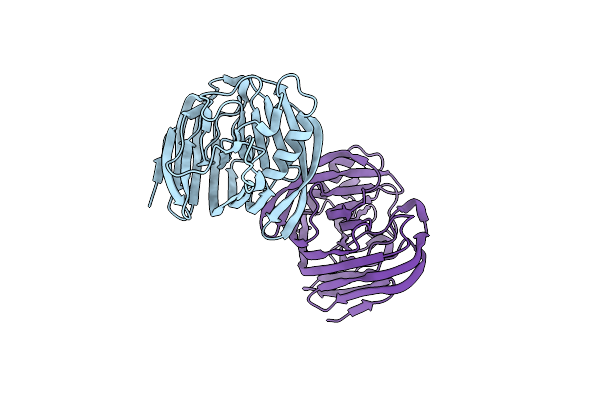 |
Use Of Synthetic Symmetrization In The Crystallization And Structure Determination Of Cela From Thermotoga Maritima
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2010-12-01 Classification: HYDROLASE Ligands: BR |
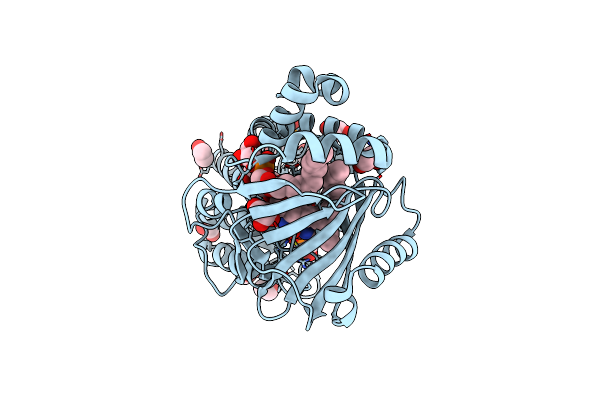 |
Crystal Structure Of A Geranylgeranyl Bacteriochlorophyll Reductase-Like (Ta0516) From Thermoplasma Acidophilum At 1.60 A Resolution
Organism: Thermoplasma acidophilum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-10-27 Classification: Flavoprotein, oxidoreductase Ligands: FAD, OZ2, EDO, GOL |
 |
Crystal Structure Of An Imelysin Peptidase (Bacova_03801) From Bacteroides Ovatus Atcc 8483 At 1.25 A Resolution
Organism: Bacteroides ovatus atcc 8483
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2010-10-20 Classification: HYDROLASE Ligands: UNL, CL, GOL |
 |
Crystal Structure Of A Protein With Unknown Function From Duf2233 Family (Bacova_00430) From Bacteroides Ovatus At 1.80 A Resolution
Organism: Bacteroides ovatus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-09-08 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: CL, SO4, EDO |
 |
Crystal Structure Of An Imelysin Peptidase (Bacova_03801) From Bacteroides Ovatus At 1.44 A Resolution
Organism: Bacteroides ovatus
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2010-07-28 Classification: HYDROLASE Ligands: MG, CL, EDO |
 |
Crystal Structure Of A Putative Ntp Pyrophosphohydrolase (Exig_1061) From Exiguobacterium Sp. 255-15 At 1.78 A Resolution
Organism: Exiguobacterium sibiricum 255-15
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2010-07-21 Classification: HYDROLASE Ligands: EDO |
 |
Crystal Structure Of Protein With Unknown Function From Duf364 Family (Zp_00559375.1) From Desulfitobacterium Hafniense Dcb-2 At 2.01 A Resolution
Organism: Desulfitobacterium hafniense
Method: X-RAY DIFFRACTION Release Date: 2010-02-02 Classification: Adenosyl binding protein Ligands: CL, IMD, EDO |
 |
Crystal Structure Of Putative Sufu (Suppressor Of Fused Protein) Homolog (Yp_208451.1) From Neisseria Gonorrhoeae Fa 1090 At 1.40 A Resolution
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2010-01-26 Classification: PROTEIN BINDING Ligands: SO4, GOL |
 |
Crystal Structure Of Cysteine Peptidase (Np_982244.1) From Bacillus Cereus Atcc 10987 At 2.50 A Resolution
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-12-15 Classification: HYDROLASE Ligands: LYS, CL |
 |
Crystal Structure Of Putative Cell Invasion Protein With Mac/Perforin Domain (Np_812351.1) From Bacteriodes Thetaiotaomicron Vpi-5482 At 2.46 A Resolution
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2009-11-24 Classification: CELL INVASION Ligands: CL, EDO, MPD |
 |
Crystal Structure Of A Putative Metal-Dependent Hydrolase (Yp_001336084.1) From Klebsiella Pneumoniae Subsp. Pneumoniae Mgh 78578 At 1.95 A Resolution
Organism: Klebsiella pneumoniae subsp. pneumoniae mgh 78578
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2009-11-10 Classification: HYDROLASE Ligands: ZN, CL, EDO |
 |
Crystal Structure Of Protein With Unknown Function From Duf35 Family (13815350) From Sulfolobus Solfataricus At 1.80 A Resolution
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-09-01 Classification: acyl-CoA binding protein Ligands: ZN, SO4, ACY |
 |
Crystal Structure Of Pleckstrin Homology Domain (Yp_926556.1) From Shewanella Amazonensis Sb2B At 1.99 A Resolution
Organism: Shewanella amazonensis sb2b
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2009-06-23 Classification: structural genomics, unknown function Ligands: GOL, PEG |
 |
Crystal Structure Of A Murein Peptide Ligase Mpl (Psyc_0032) From Psychrobacter Arcticus 273-4 At 1.65 A Resolution
Organism: Psychrobacter arcticus 273-4
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2009-06-16 Classification: LIGASE |
 |
Crystal Structure Of A Putative Glycoside Hydrolase (Bt_2081) From Bacteroides Thetaiotaomicron Vpi-5482 At 2.05 A Resolution
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2009-05-26 Classification: Ca-BINDING PROTEIN Ligands: CA, NA, CAC, ACT, PEG, P6G |
 |
Crystal Structure Of A Nlpc/P60 Family Protein (Bce_2878) From Bacillus Cereus Atcc 10987 At 1.79 A Resolution
Organism: Bacillus cereus atcc 10987
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2009-05-05 Classification: HYDROLASE Ligands: ALA, DGL, PO4, PG4 |
 |
Crystal Structure Of A Tetracenomycin Polyketide Synthesis Protein (Tcmj) From Xanthomonas Campestris Pv. Campestris At 1.60 A Resolution
Organism: Xanthomonas campestris pv. campestris
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2009-05-05 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN, ACT |
 |
Crystal Structure Of A Duf1470 Family Protein (Jann_2411) From Jannaschia Sp. Ccs1 At 1.45 A Resolution
Organism: Jannaschia sp. ccs1
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2009-04-21 Classification: METAL BINDING PROTEIN Ligands: ZN, NI, NA, GOL, ACT |

