Search Count: 37
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: SIGNALING PROTEIN |
 |
Organism: Latrodectus tredecimguttatus
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: TOXIN |
 |
Organism: Latrodectus tredecimguttatus
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: TOXIN |
 |
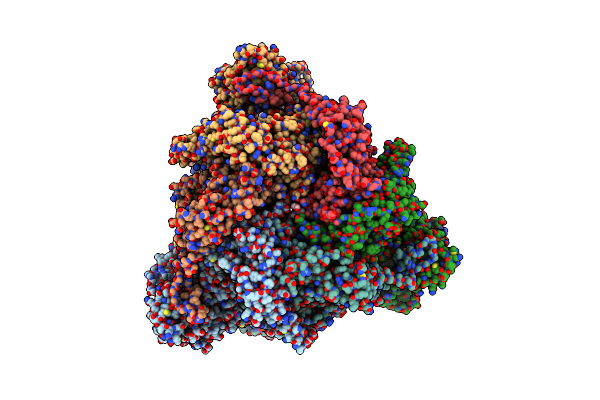
Structure Of The Peroxisomal Pex1/Pex6 Atpase Complex Bound To A Substrate In Single Seam State
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-10-04 Classification: TRANSLOCASE Ligands: ATP, MG, ADP |
 |
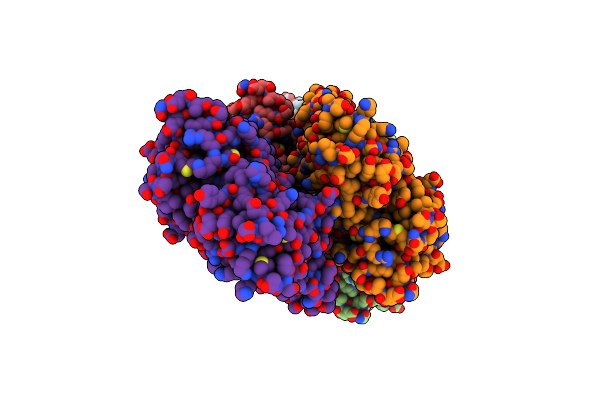
Structure Of The Peroxisomal Pex1/Pex6 Atpase Complex Bound To A Substrate In Twin Seam State
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-10-04 Classification: TRANSLOCASE Ligands: ATP, MG, ADP |
 |
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-10 Classification: STRUCTURAL PROTEIN Ligands: ADP, MG, BEF |
 |
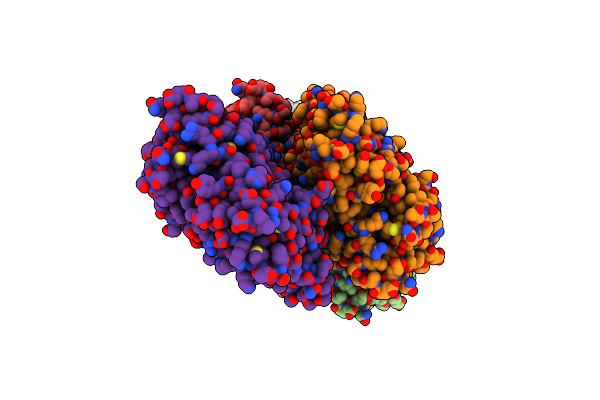
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-10 Classification: STRUCTURAL PROTEIN Ligands: ADP, MG, PO4 |
 |
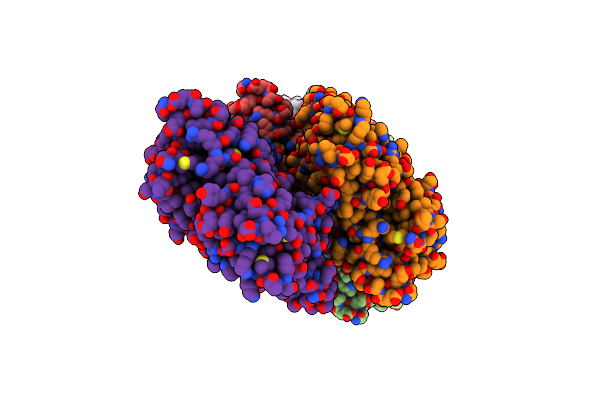
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-10 Classification: STRUCTURAL PROTEIN Ligands: ADP, MG |
 |
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-10 Classification: STRUCTURAL PROTEIN Ligands: ADP, CA, BEF |
 |
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-10 Classification: STRUCTURAL PROTEIN Ligands: ADP, CA, PO4 |
 |
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-10 Classification: STRUCTURAL PROTEIN Ligands: ADP, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-06-29 Classification: STRUCTURAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-06-29 Classification: RNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: RNA BINDING PROTEIN |
 |
Organism: Chaetomium thermophilum
Method: ELECTRON MICROSCOPY Release Date: 2022-02-09 Classification: ENDOCYTOSIS |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-02-19 Classification: PROTEIN TRANSPORT |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-01-29 Classification: PROTEIN TRANSPORT |
 |
The X-Ray Structure Of The Sam-Dependent Uroporphyrinogen Iii Methyltransferase Nire From Pseudomonas Aeruginosa In Complex With Sah
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-06-01 Classification: TRANSFERASE Ligands: SAH |
 |
The X-Ray Structure Of The Sam-Dependent Uroporphyrinogen Iii Methyltransferase Nire From Pseudomonas Aeruginosa In Complex With Sah And Uroporphyrinogen Iii
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-06-01 Classification: TRANSFERASE Ligands: SAH, UP2 |
 |
Crystal Structure Of The Sulfane Dehydrogenase Soxcd From Paracoccus Pantotrophus
Organism: Paracoccus pantotrophus
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2010-12-08 Classification: OXIDOREDUCTASE/ELECTRON TRANSPORT Ligands: MTE, 2MO, CO, GOL, HEC, CA |

