Search Count: 20
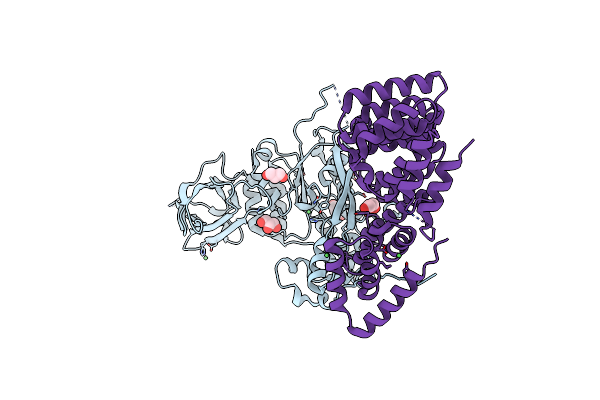 |
Structure Of The Pseudomonas Putida Algkx Modification And Secretion Complex
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2022-12-14 Classification: TRANSFERASE Ligands: NI, CL, GOL |
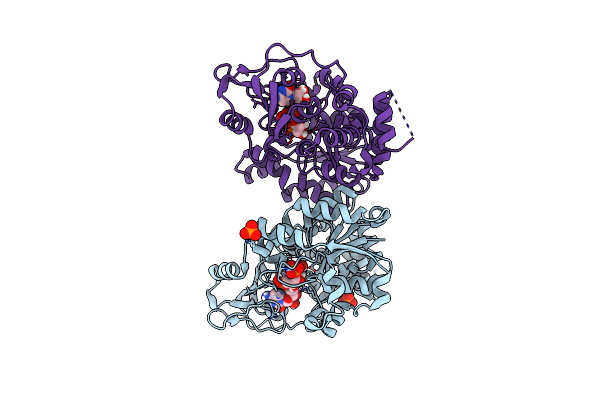 |
Organism: Raoultella terrigena
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-11-09 Classification: TRANSFERASE Ligands: PO4, C5P, CMK |
 |
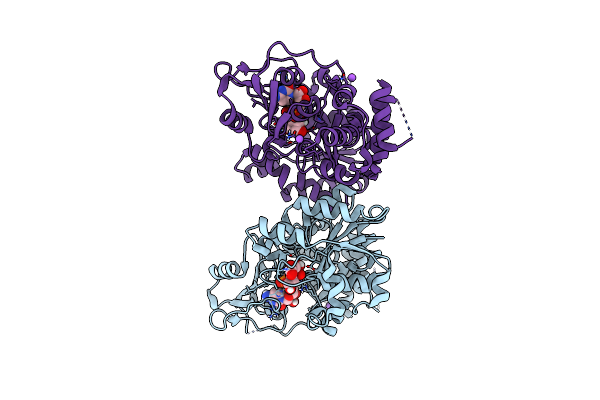
Organism: Raoultella terrigena
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-11-09 Classification: TRANSFERASE Ligands: C5P, KDO, NA, CL |
 |
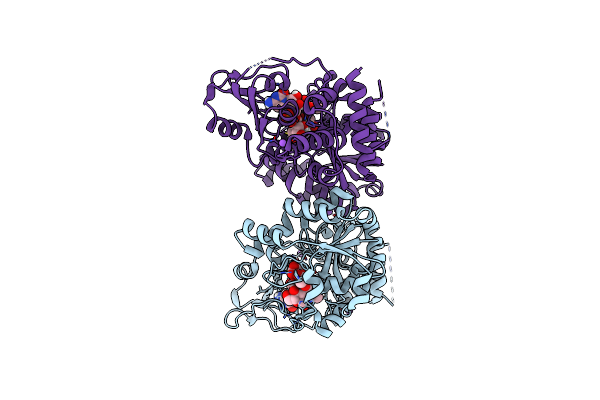
Organism: Raoultella terrigena
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-11-09 Classification: TRANSFERASE Ligands: C5P, KDO, CL, NA |
 |
Organism: Raoultella terrigena
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-11-09 Classification: TRANSFERASE Ligands: PXV, C5P, NA |
 |
Organism: Raoultella terrigena
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-11-09 Classification: TRANSFERASE Ligands: C5P, NA, KDO |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2022-08-03 Classification: STRUCTURAL PROTEIN Ligands: MG, CL |
 |
Organism: Neosartorya fumigata (strain atcc mya-4609 / af293 / cbs 101355 / fgsc a1100)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-02-12 Classification: HYDROLASE Ligands: NAG, MAN, CL, ZN |
 |
Cs-Rosetta Determined Structures Of The N-Terminal Domain Of Algf From P. Aeruginosa
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: SOLUTION NMR Release Date: 2019-04-17 Classification: CELL ADHESION |
 |
Cs-Rosetta Determined Structures Of The C-Terminal Domain Of Algf From P. Aeruginosa
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: SOLUTION NMR, SOLUTION SCATTERING Release Date: 2019-04-17 Classification: CELL ADHESION |
 |
Organism: Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2019-01-02 Classification: PROTEIN BINDING Ligands: CO, SO4, CL, SCN |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2014-10-01 Classification: TRANSFERASE |
 |
Organism: Pseudomonas syringae pv. tomato
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-01-15 Classification: ISOMERASE |
 |
Organism: Pseudomonas syringae pv. tomato
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2014-01-15 Classification: ISOMERASE |
 |
Crystal Structure Of Clostridium Difficile Toxin A Fragment Tcda-A1 Bound To A20.1 Vhh
Organism: Clostridium difficile, Lama glama
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2013-12-11 Classification: IMMUNE SYSTEM |
 |
Organism: Clostridium difficile, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2013-12-11 Classification: IMMUNE SYSTEM |
 |
Organism: Clostridium difficile, Lama glama
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2013-12-11 Classification: IMMUNE SYSTEM |
 |
Organism: Clostridium difficile, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-12-11 Classification: IMMUNE SYSTEM |
 |
Organism: Clostridium difficile, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2013-12-11 Classification: IMMUNE SYSTEM |
 |
Organism: Clostridium difficile, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-12-11 Classification: IMMUNE SYSTEM |

