Search Count: 15
 |
Crystal Structure Of The Sars-Cov-2 (Covid-19) Main Protease In Complex With Uaw243
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-06-24 Classification: HYDROLASE Ligands: GOL |
 |
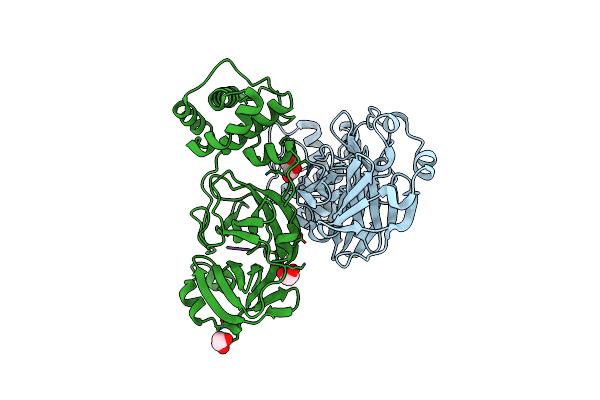
Crystal Structure Of The Sars-Cov-2 (Covid-19) Main Protease In Complex With Uaw241
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-06-17 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GOL |
 |
Crystal Structure Of The Sars-Cov-2 (Covid-19) Main Protease In Complex With Inhibitor Uaw246
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2020-06-17 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GOL, NA |
 |
Crystal Structure Of The Sars-Cov-2 (Covid-19) Main Protease In Complex With Inhibitor Uaw247
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-06-17 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GOL, NA |
 |
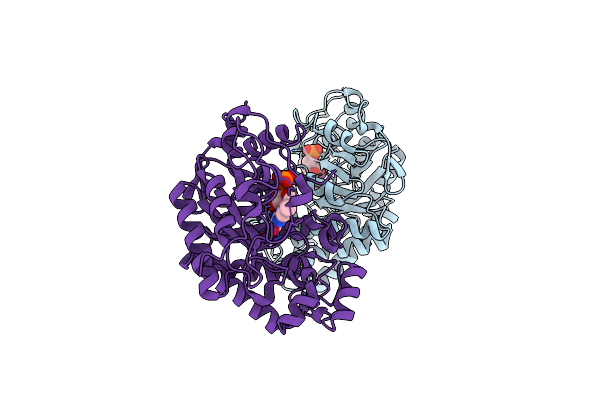
Crystal Structure Of The Sars-Cov-2 (Covid-19) Main Protease In Complex With Inhibitor Uaw248
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-06-17 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GOL, NA, DMS |
 |
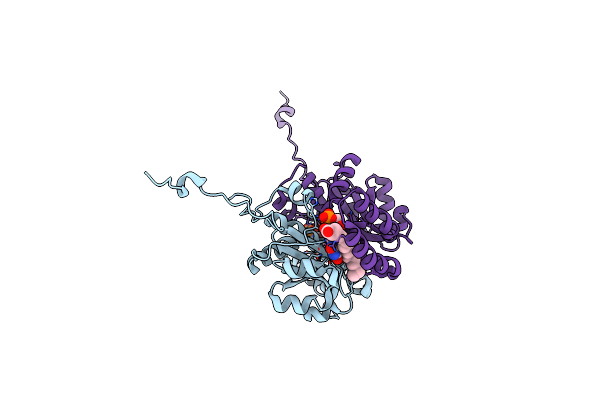
Organism: Pichia
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-06-26 Classification: OXIDOREDUCTASE Ligands: FMN |
 |
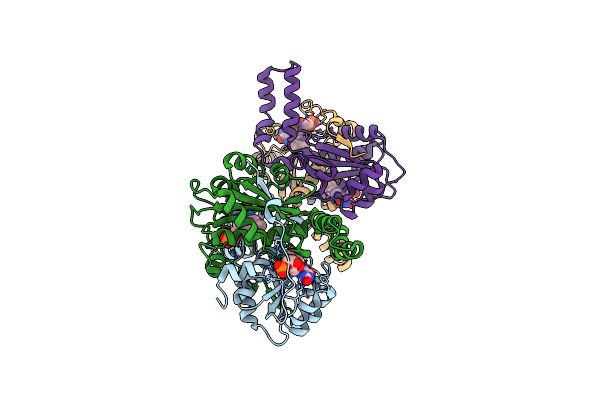
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2015-02-25 Classification: OXIDOREDUCTASE Ligands: FMN, KTC, CL |
 |
Crystal Structure Of Cla-Er, A Novel Enone Reductase Catalyzing A Key Step Of A Gut-Bacterial Fatty Acid Saturation Metabolism, Biohydrogenation
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2015-02-25 Classification: OXIDOREDUCTASE Ligands: FMN |
 |
Organism: Kluyveromyces marxianus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-02-11 Classification: FLAVOPROTEIN Ligands: FMN |
 |
Crystal Structure Of Old Yellow Enzyme From Candida Macedoniensis Aku4588 Complexed With P-Hydroxybenzaldehyde
Organism: Kluyveromyces marxianus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-02-11 Classification: FLAVOPROTEIN Ligands: FMN, HBA |
 |
Organism: Aeromonas jandaei
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-07-09 Classification: LYASE Ligands: PLG, GLY |
 |
Organism: Aeromonas jandaei
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-07-09 Classification: LYASE Ligands: PLG |
 |
Crystal Structure Of Conjugated Polyketone Reductase C2 From Candida Parapsilosis
Organism: Candida parapsilosis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-08-07 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Conjugated Polyketone Reductase C2 From Candida Parapsilosis Complexed With Nadph
Organism: Candida parapsilosis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-08-07 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
Crystal Structure Of Stam2 Sh3 Domain In Complex With A Ubpy-Derived Peptide
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2003-12-23 Classification: SIGNALING PROTEIN/SIGNALING PROTEIN Ligands: PO4 |

