Search Count: 34
 |
Crystal Structure Of Cdk2 In Complex With Cyclin A Inhibitor 6-[(E)-2-(4-Chlorophenyl)Ethenyl]-2-{[(2R)-3-(4-Hydroxyphenyl)-1-Methoxy-1-Oxopropan-2-Yl]Carbamoyl}Quinoline-4-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-04-05 Classification: TRANSFERASE/INHIBITOR Ligands: OW6 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2022-12-28 Classification: COMPLEX (ISOMERASE/KINASE) Ligands: XYU, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.07 Å Release Date: 2022-12-28 Classification: COMPLEX (ISOMERASE/KINASE) Ligands: XZ3, CL |
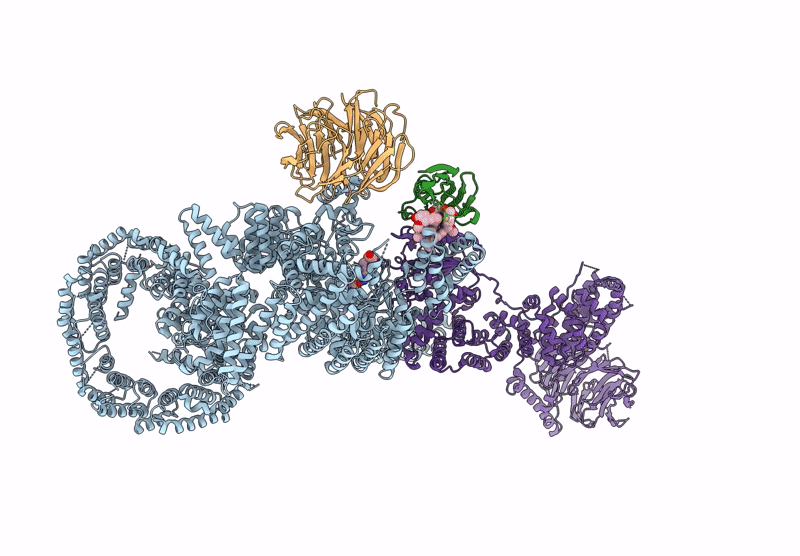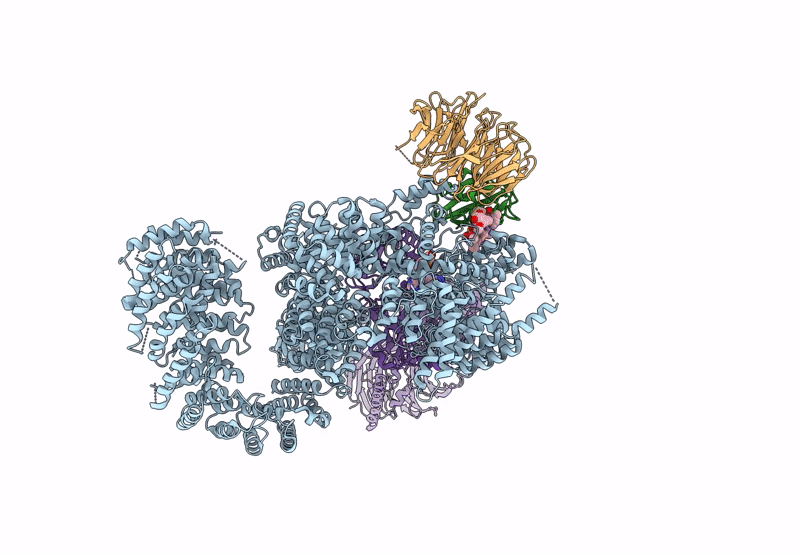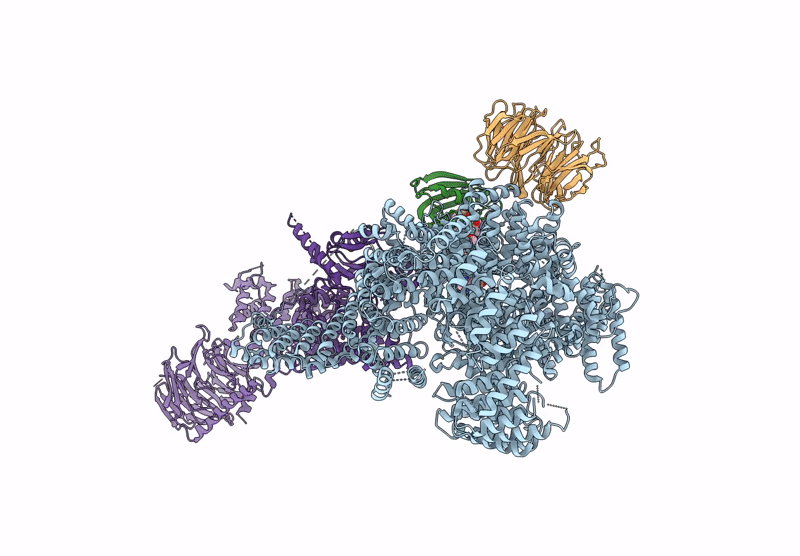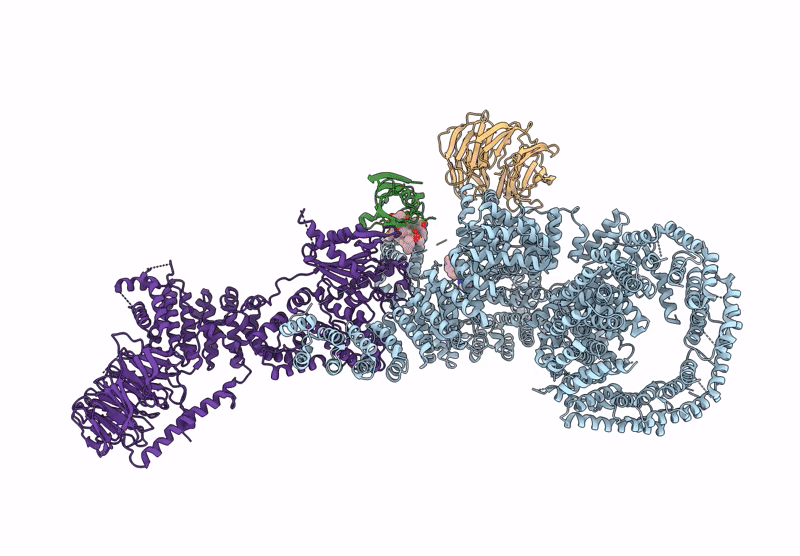 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-12-28 Classification: COMPLEX (ISOMERASE/KINASE) Ligands: XZ9, XYU |
 |
Organism: Streptomyces scabies (strain 87.22)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2016-06-22 Classification: OXIDOREDUCTASE Ligands: HEM, TRP, GOL, CL |
 |
Organism: Streptomyces scabies (strain 87.22)
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2016-06-22 Classification: OXIDOREDUCTASE Ligands: HEM, TRP, GOL, CL |
 |
Directed Evolution And Rational Design Of A De Novo Designed Esterase Toward Improved Catalysis. Northeast Structural Genomics Consortium (Nesg) Target Or184
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2013-05-22 Classification: Structural Genomics, Unknown Function Ligands: BR, GOL, PE5 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2012-06-27 Classification: LYASE Ligands: SO4, NA |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-06-06 Classification: LYASE Ligands: PO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-06-06 Classification: LYASE/LYASE INHIBITOR Ligands: 0CU, PO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2012-06-06 Classification: LYASE Ligands: SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-06-06 Classification: LYASE Ligands: PO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-06-06 Classification: LYASE/LYASE INHIBITOR Ligands: 0CU, PO4, NA |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2012-06-06 Classification: LYASE/LYASE INHIBITOR Ligands: 0CT, PO4 |
 |
Crystal Structure Of Engineered Protein, Northeast Structural Genomics Consortium Target Or120
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-02-08 Classification: ISOMERASE |
 |
X-Ray Structure Of De Novo Design Cysteine Esterase Fr29, Northeast Structural Genomics Consortium Target Or52
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2011-12-07 Classification: Structural Genomics, Unknown Function |
 |
Crystal Structure Of De Novo Designed Cysteine Esterase Ech14, Northeast Structural Genomics Consortium Target Or54
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.23 Å Release Date: 2011-12-07 Classification: TRANSFERASE |
 |
Crystal Structure Of De Novo Design Of Cystein Esterase Ech13 At The Resolution 1.6A, Northeast Structural Genomics Consortium Target Or51
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2011-11-23 Classification: HYDROLASE Ligands: PO4, MG, PEG, NA |
 |
Crystal Structure Of Engineered Protein. Northeast Structural Genomics Consortium Target Or48
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2011-11-23 Classification: Structural Genomics, Unknown Function |
 |
Three Dimensional Structure Of De Novo Designed Cysteine Esterase Ech19, Northeast Structural Genomics Consortium Target Or49
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2011-10-26 Classification: HYDROLASE Ligands: SO4, NA |

