Search Count: 18
 |
Solution Nmr Structure Of The Bound Antimicrobial Peptide Kg18 To Pseudomonas Aeruginosa Lps Bicelle At Ph 4
Organism: Dengue virus
Method: SOLUTION NMR Release Date: 2025-09-24 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-09-24 Classification: ANTIMICROBIAL PROTEIN |
 |
Solution Nmr Structure Of The Bound Antimicrobial Peptide Vr18 To Pseudomonas Aeruginosa Lps Bicelle At Ph 4
Organism: Dengue virus
Method: SOLUTION NMR Release Date: 2025-09-17 Classification: ANTIMICROBIAL PROTEIN |
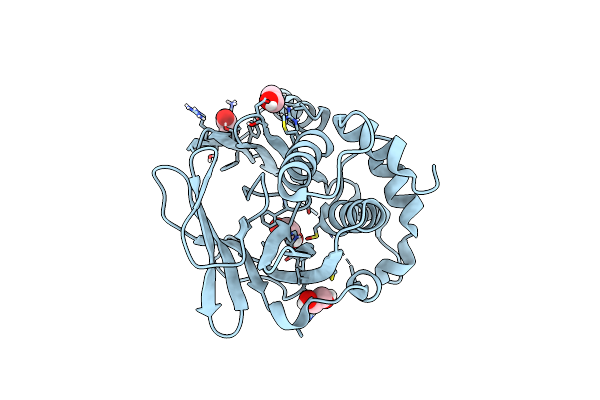 |
Organism: Streptomyces hygroscopicus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-06-12 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO, ACT |
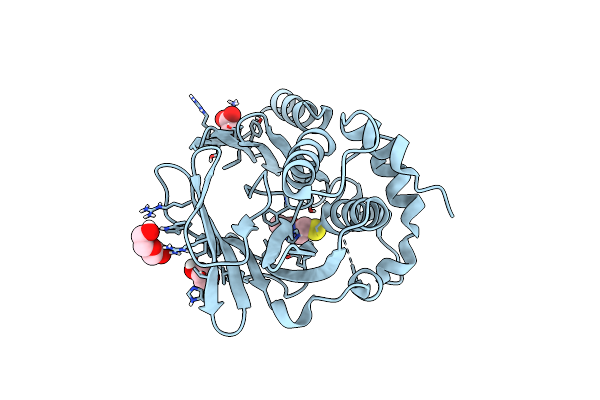 |
Organism: Streptomyces hygroscopicus
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2024-05-01 Classification: BIOSYNTHETIC PROTEIN Ligands: W0K, PEG, ACT, EDO |
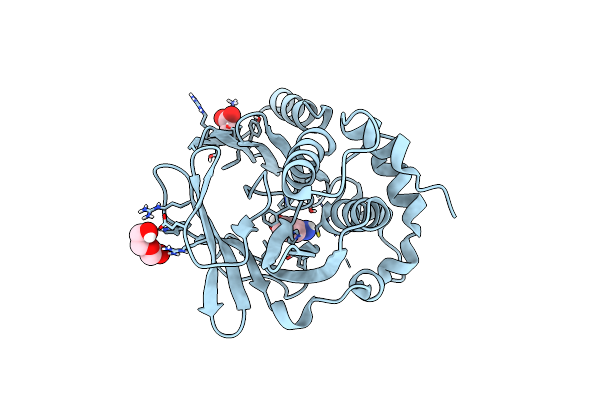 |
Organism: Streptomyces hygroscopicus
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2024-05-01 Classification: BIOSYNTHETIC PROTEIN Ligands: K5V, PEG, ACT |
 |
Organism: Streptomyces hygroscopicus
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2024-05-01 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO, ACT, W0E, SO4, PEG |
 |
Organism: Streptomyces hygroscopicus
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2024-04-17 Classification: BIOSYNTHETIC PROTEIN Ligands: PNY, ACT |
 |
Cryo-Em Structure Of The Mycobacterium Tuberculosis Cytochrome Bcc:Aa3 Supercomplex And A Novel Inhibitor Targeting Subunit Cytochrome Ci
Organism: Mycobacterium tuberculosis variant bovis bcg
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: OXIDOREDUCTASE Ligands: FES, HEM, HEC, HEA |
 |
Solution Structure Of Subunit Epsilon Of The Mycobacterium Abscessus F-Atp Synthase
Organism: Mycobacteroides abscessus
Method: SOLUTION NMR Release Date: 2023-03-08 Classification: ELECTRON TRANSPORT |
 |
Solution Structure Of Subunit Epsilon Of The Mycobacterium Abscessus F-Atp Synthase
Organism: Mycobacteroides abscessus
Method: SOLUTION NMR Release Date: 2022-10-05 Classification: BIOSYNTHETIC PROTEIN |
 |
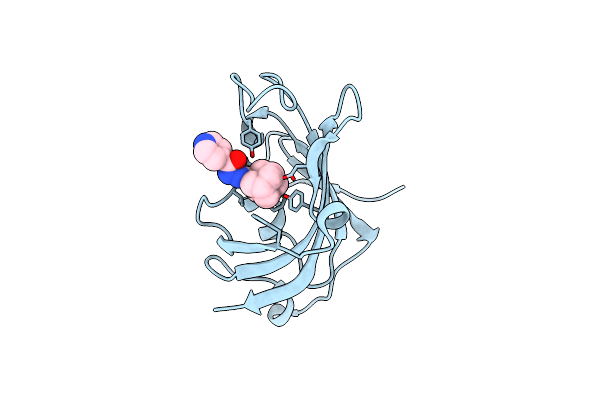
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-02-12 Classification: ISOMERASE Ligands: IMD, CL |
 |
Crystal Structure Of Fk506 Binding Domain Of Plasmodium Vivax Fkbp35 In Complex With Inhibitor D5
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2013-12-04 Classification: ISOMERASE Ligands: D5I |
 |
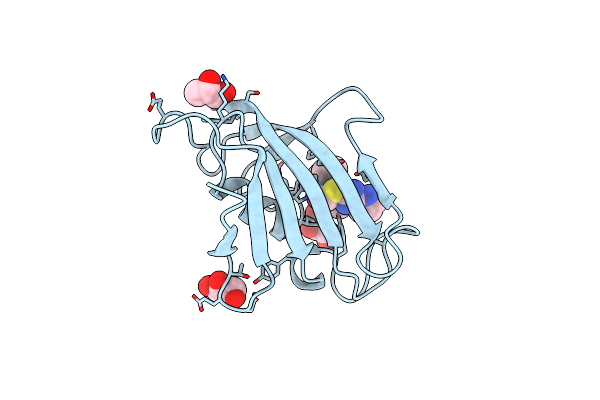
Crystal Structure Of Fk506 Binding Domain Of Plasmodium Falciparum Fkbp35 In Complex With D44
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2013-09-11 Classification: ISOMERASE/ISOMERASE INHIBITOR Ligands: D44 |
 |
Crystal Structure Of Fk506 Binding Domain Of Plasmodium Vivax Fkbp35 In Complex With D44
Organism: Plasmodium vivax sai-1
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2013-09-11 Classification: ISOMERASE/ISOMERASE INHIBITOR Ligands: D44, GOL |
 |
 |
 |
Plasmodium Falciparum Putative Fk506-Binding Protein Pfl2275C, C-Terminal Tpr-Containing Domain
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2006-01-24 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |

