Search Count: 39
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: TRANSCRIPTION |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: TRANSCRIPTION |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE Ligands: CA |
 |
Klebsiella Pneumoniae Maltohexaose-Producing Alpha-Amylase In Complex With Acarbose
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE Ligands: CA |
 |
Domain N Deletion Mutant Of Klebsiella Pneumoniae Maltohexaose-Producing Alpha-Amylase In Complex With Maltohexaose
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE Ligands: CA |
 |
Klebsiella Pneumoniae Maltohexaose-Producing Alpha-Amylase, Terbium Derivative
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE Ligands: CA, TB |
 |
Crystal Structure Of The S103F Mutant Of Bacillus Subtilis (Natto) Yabj Protein.
Organism: Bacillus subtilis subsp. natto (strain best195)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2021-03-03 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of The S103F Mutant Of Bacillus Subtilis (Natto) Yabj Protein.
Organism: Bacillus subtilis subsp. natto (strain best195)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-03-03 Classification: UNKNOWN FUNCTION Ligands: SO4, GOL |
 |
Crystal Structure Of The S103F Mutant Of Bacillus Subtilis (Natto) Yabj Protein.
Organism: Bacillus subtilis subsp. natto (strain best195)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-03-03 Classification: UNKNOWN FUNCTION Ligands: ZN, CL, BTB, MG |
 |
Crystal Structure Of Glycoside Hydrolase Family 11 Beta-Xylanase From Streptomyces Olivaceoviridis E-86
Organism: Streptomyces olivaceoviridis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-12-30 Classification: HYDROLASE Ligands: CL |
 |
Crystal Structure Of Glycoside Hydrolase Family 11 Beta-Xylanase From Streptomyces Olivaceoviridis E-86 In Complex With Alpha-L-Arabinofuranosyl Xylotetraose
Organism: Streptomyces olivaceoviridis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-12-30 Classification: HYDROLASE Ligands: CL, NA |
 |
Crystal Structure Of Glycoside Hydrolase Family 11 Beta-Xylanase From Streptomyces Olivaceoviridis E-86 In Complex With 4-O-Methyl-Alpha-D-Glucuronopyranosyl Xylotetraose
Organism: Streptomyces olivaceoviridis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-12-30 Classification: HYDROLASE Ligands: CL, NA |
 |
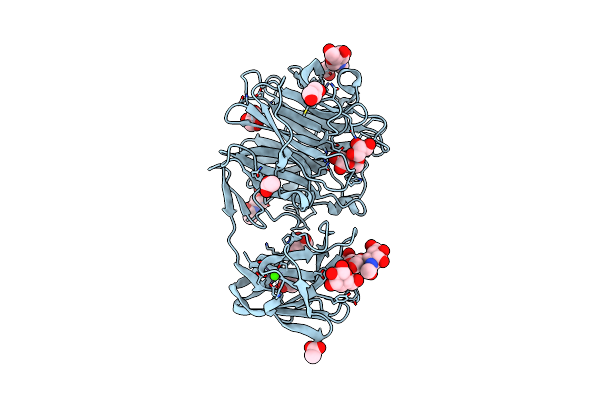
Crystal Structure Of Exo-Beta-1,3-Galactanase From Phanerochaete Chrysosporium Pc1,3Gal43A Apo Form
Organism: Phanerochaete chrysosporium
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-11-04 Classification: HYDROLASE Ligands: NAG, CA, CIT |
 |
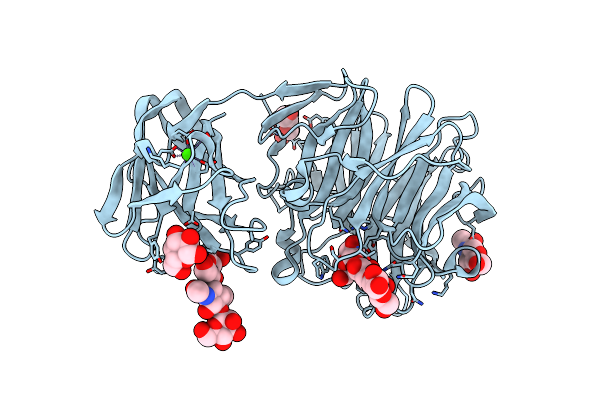
Crystal Structure Of Exo-Beta-1,3-Galactanase From Phanerochaete Chrysosporium Pc1,3Gal43A With Galactose
Organism: Phanerochaete chrysosporium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-11-04 Classification: HYDROLASE Ligands: NAG, CA, GLA, GAL, ACY, ACT, GOL |
 |
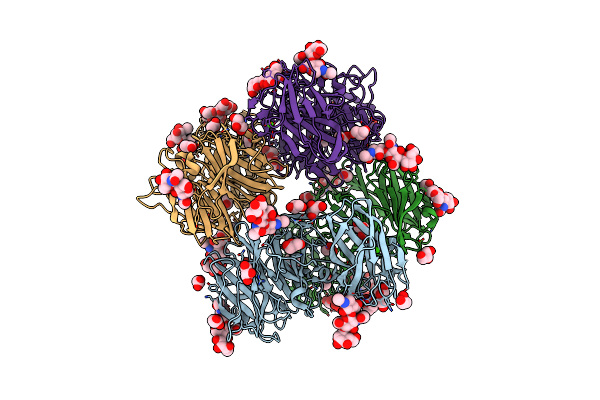
Crystal Structure Of Exo-Beta-1,3-Galactanase From Phanerochaete Chrysosporium Pc1,3Gal43A E208Q With Beta-1,3-Galactotriose
Organism: Phanerochaete chrysosporium
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-11-04 Classification: HYDROLASE Ligands: NAG, CA, GAL |
 |
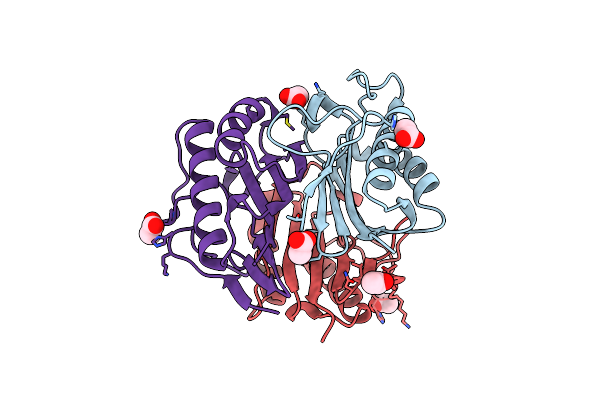
Crystal Structure Of Exo-Beta-1,3-Galactanase From Phanerochaete Chrysosporium Pc1,3Gal43A E208A With Beta-1,3-Galactotriose
Organism: Phanerochaete chrysosporium
Method: X-RAY DIFFRACTION Release Date: 2020-11-04 Classification: HYDROLASE Ligands: CA, ACT, GOL, NAG |
 |
Crystal Structure Of Wild-Type Yabj Protein From Bacillus Subtilis (Natto).
Organism: Bacillus subtilis subsp. natto (strain best195)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-08-15 Classification: UNKNOWN FUNCTION Ligands: ACY |
 |
Organism: Paenibacillus sp. 598k
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-07-26 Classification: HYDROLASE, TRANSFERASE Ligands: CA, NI, MG, SO4, MES, EDO |
 |
Crystal Structure Of Paenibacillus Sp. 598K Alpha-1,6-Glucosyltransferase Complexed With Acarbose
Organism: Paenibacillus sp. 598k
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-07-26 Classification: HYDROLASE, TRANSFERASE Ligands: CA, NI, MG, SO4, MES, EDO |
 |
Crystal Structure Of Paenibacillus Sp. 598K Alpha-1,6-Glucosyltransferase Complexed With Maltohexaose
Organism: Paenibacillus sp. 598k
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-07-26 Classification: HYDROLASE, TRANSFERASE Ligands: CA, BGC, GLC, NI, MG, SO4, MES, EDO |

