Search Count: 13
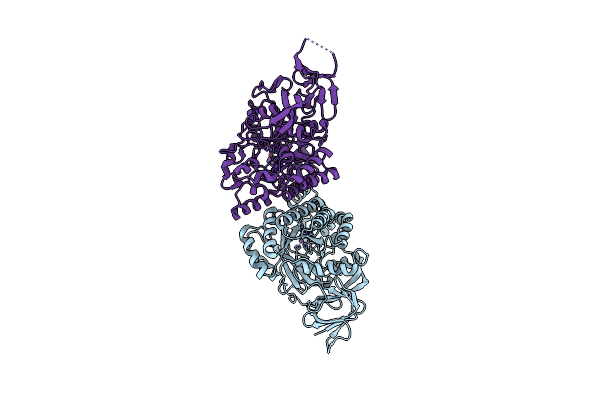 |
2.8 Angstrom Resolution Crystal Structure Of D-Hydantoinase From Bacillus Sp. Ar9 In C2221 Space Group
Organism: Bacillus sp. ar9
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-05-21 Classification: HYDROLASE Ligands: MN |
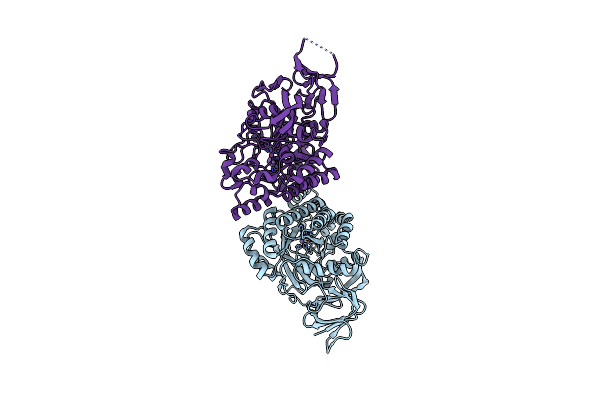 |
Crystal Structure Of D-Hydantoinase From Bacillus Sp. Ar9 In C2221 Space Group
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-05-07 Classification: HYDROLASE Ligands: MN |
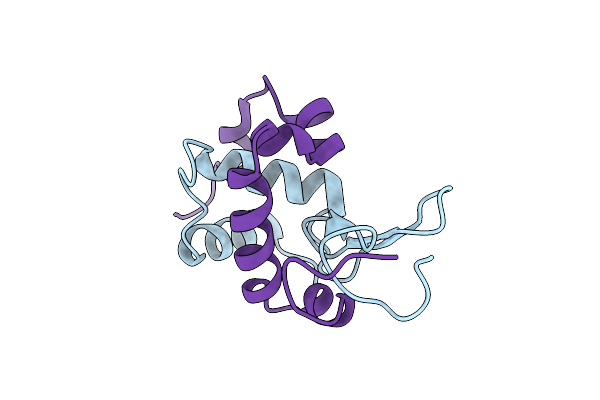 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-10-19 Classification: HYDROLASE |
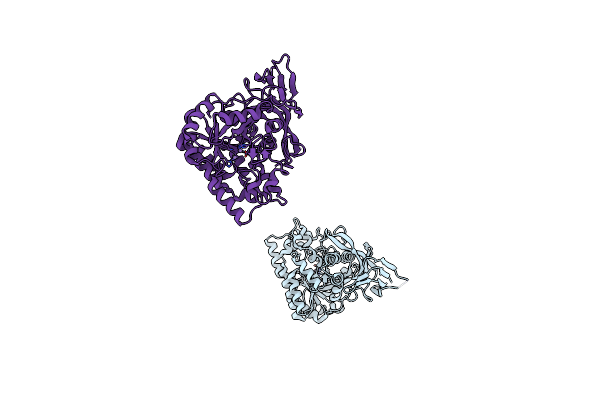 |
Molecular Structure Of D-Hydantoinase From A Bacillus Sp. Ar9: Evidence For Mercury Inhibition
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2005-03-01 Classification: HYDROLASE Ligands: MN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2001-05-09 Classification: HORMONE/GROWTH FACTOR |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2001-05-09 Classification: HORMONE/GROWTH FACTOR |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1998-07-08 Classification: SIGNAL TRANSDUCTION |
 |
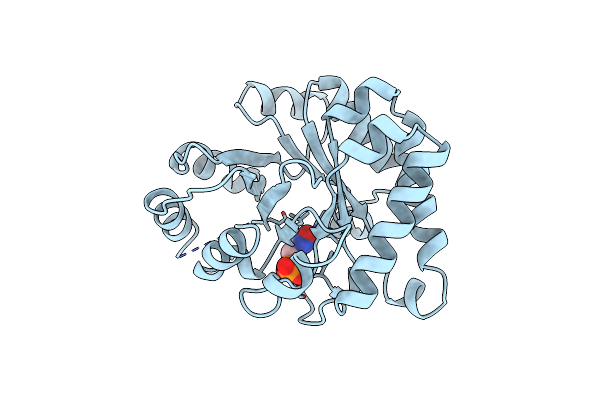
Three New Crystal Structures Of Point Mutation Variants Of Monotim: Conformational Flexibility Of Loop-1,Loop-4 And Loop-8
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1995-10-15 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: PGA |
 |
Three New Crystal Structures Of Point Mutation Variants Of Monotim: Conformational Flexibility Of Loop-1,Loop-4 And Loop-8
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1995-09-15 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: PGH |
 |
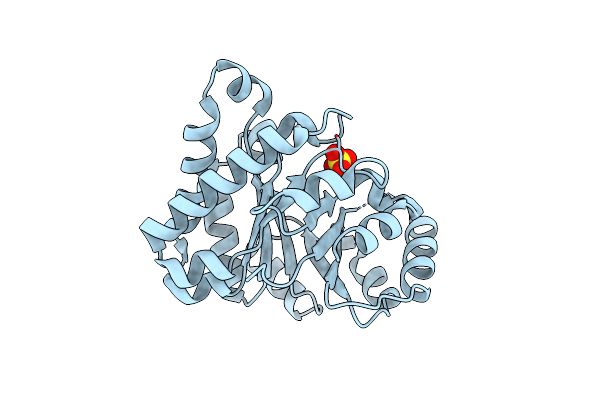
Large Scale Structural Rearrangements Of The Front Loops In Monomerised Triosephosphate Isomerase, As Deduced From The Comparison Of The Structural Properties Of Monotim And Its Point Mutation Variant Monoss
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1994-09-30 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) |
 |
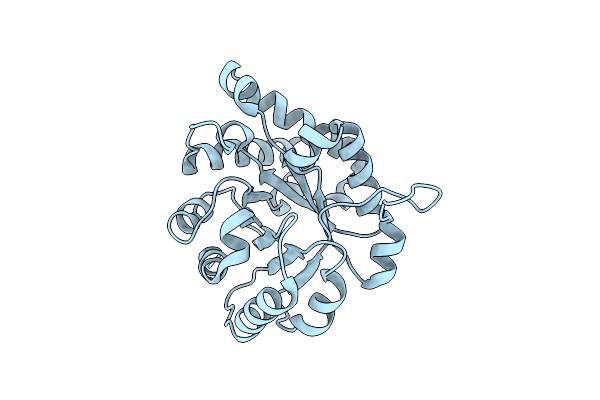
The Crystal Structure Of An Engineered Monomeric Triosephosphate Isomerase, Monotim: The Correct Modelling Of An Eight-Residue Loop
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1994-07-31 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: SO4 |
 |
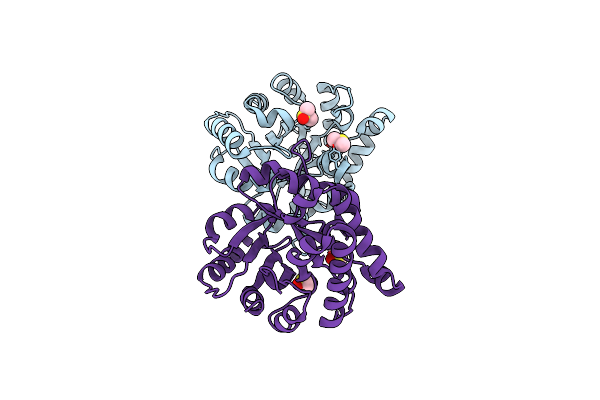
Comparison Of The Structures And The Crystal Contacts Of Trypanosomal Triosephosphate Isomerase In Four Different Crystal Forms
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1994-05-31 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) |
 |
Comparison Of The Structures And The Crystal Contacts Of Trypanosomal Triosephosphate Isomerase In Four Different Crystal Forms
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1994-05-31 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: DMS |

