Search Count: 193
 |
Crystal Structure Of The Histidine Kinase Vc2136 From Vibrio Cholerae Serotype O1
Organism: Vibrio cholerae o1 biovar el tor str. n16961
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: TRANSFERASE Ligands: SO4, CL |
 |
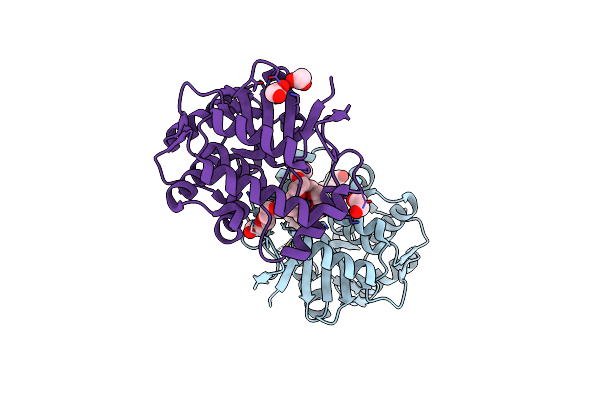
Crystal Structure Of The Sars-Cov-2 2'-O-Methyltransferase With (M7Gpppa)Pupu (Cap-0) And S-Adenosyl-L-Homocysteine (Sah).
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: TRANSFERASE, Viral Protein Ligands: SAH, CL, SO4, ZN, MGT |
 |
Crystal Structure Of The Surface Protein (Cd630_07380) From Clostridium Difficile Strain 630
Organism: Clostridioides difficile 630
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of N-Terminal Domain Of Fic Family Protein From Bordetella Bronchiseptica
Organism: Bordetella bronchiseptica
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2024-10-30 Classification: TRANSFERASE |
 |
Crystal Structure Of The Dtdp-4-Dehydrorhamnose Reductase From Streptococcus Pneumoniae.
Organism: Streptococcus pneumoniae taiwan19f-14
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2023-02-22 Classification: OXIDOREDUCTASE |
 |
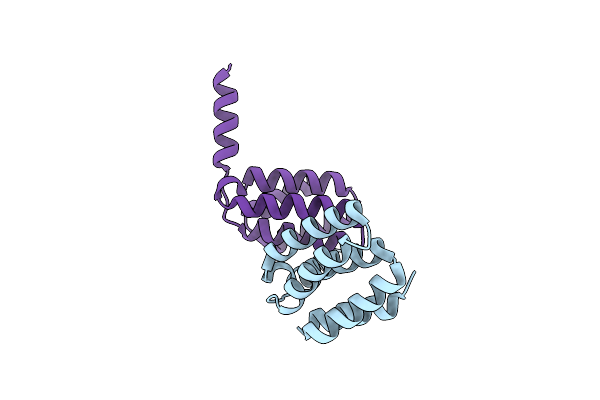
Crystal Structure Of The Threonine Synthase From Streptococcus Pneumoniae In Complex With Pyridoxal 5-Phosphate.
Organism: Streptococcus pneumoniae tigr4
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2023-02-15 Classification: LYASE Ligands: CL, NA, PLP, GOL, MLT |
 |
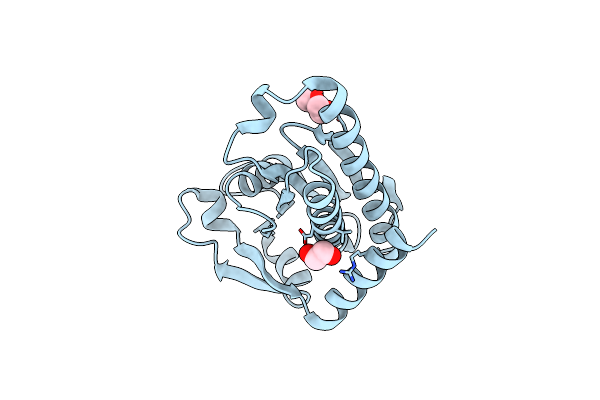
Crystal Structure Of The C-Terminal Fragment Of Aaa Atpase From Streptococcus Pneumoniae.
Organism: Streptococcus pneumoniae tigr4
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2023-02-15 Classification: HYDROLASE Ligands: CL |
 |
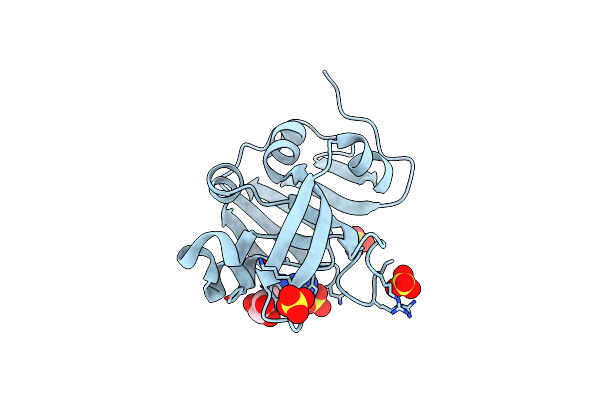
Crystal Structure Of The Hypothetical Protein (Acx60_00475) From Acinetobacter Baumannii
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-11-30 Classification: UNKNOWN FUNCTION Ligands: PEG, EDO |
 |
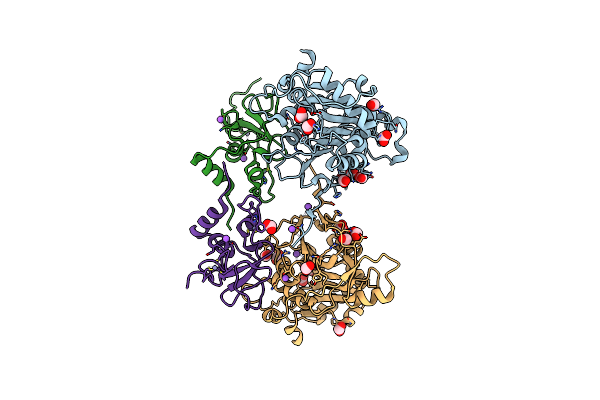
Crystal Structure Of Putative Pterin Binding Protein (Prur) From Klebsiella Pneumoniae In Complex With Neopterin
Organism: Klebsiella pneumoniae subsp. pneumoniae sa1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-08-10 Classification: PTERIN BINDING PROTEIN Ligands: NEU, CL, SO4 |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-04-13 Classification: TRANSFERASE |
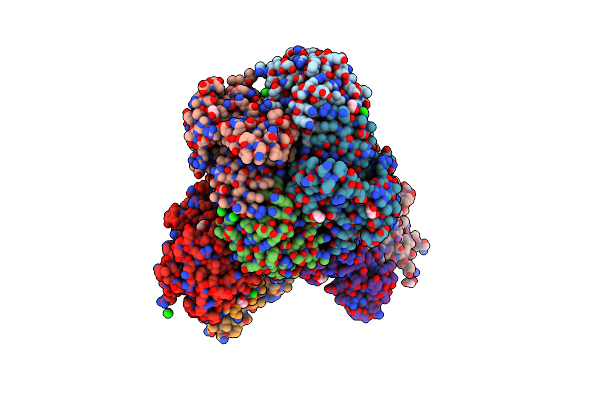 |
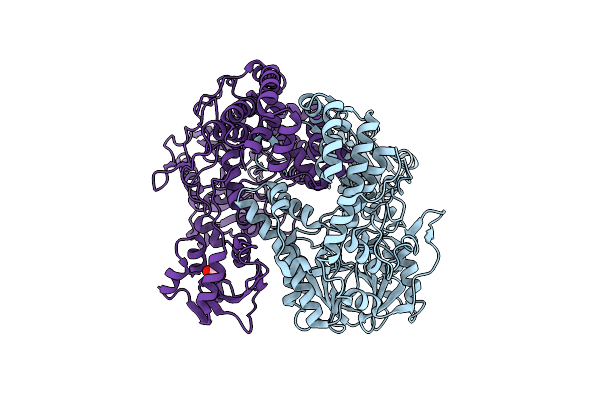
Crystal Structure Of Putataive Short-Chain Dehydrogenase/Reductase (Fabg) From Klebsiella Pneumoniae Subsp. Pneumoniae Ntuh-K2044 In Complex With Nadh
Organism: Klebsiella pneumoniae subsp. pneumoniae ntuh-k2044
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-03-02 Classification: BIOSYNTHETIC PROTEIN Ligands: K, CL, NAI, GOL, EDO |
 |
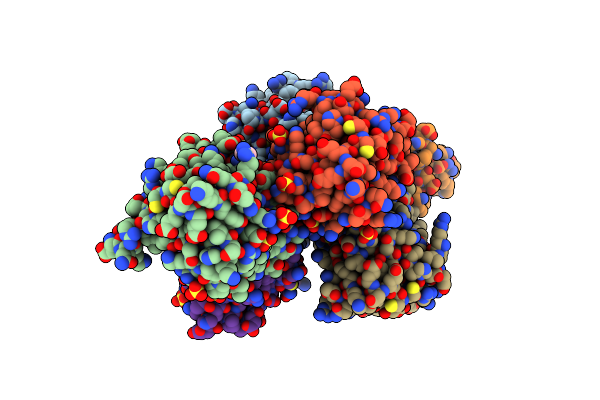
Organism: Klebsiella pneumoniae subsp. pneumoniae ntuh-k2044
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2022-02-02 Classification: HYDROLASE Ligands: EDO |
 |
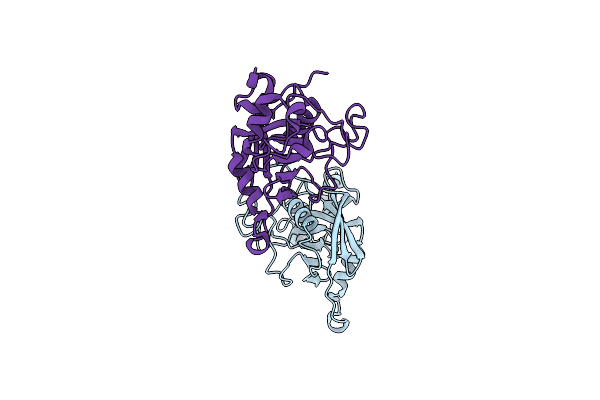
Crystal Structure Of The Dna-Binding Transcriptional Repressor Deor From Escherichia Coli Str. K-12
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-12-01 Classification: DNA BINDING PROTEIN Ligands: EDO, SO4, CL |
 |
Crystal Structure Of The Peptidyl-Prolyl Cis-Trans Isomerase (Ppib) From Streptococcus Pyogenes.
Organism: Streptococcus pyogenes mgas5005
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2021-12-01 Classification: ISOMERASE |
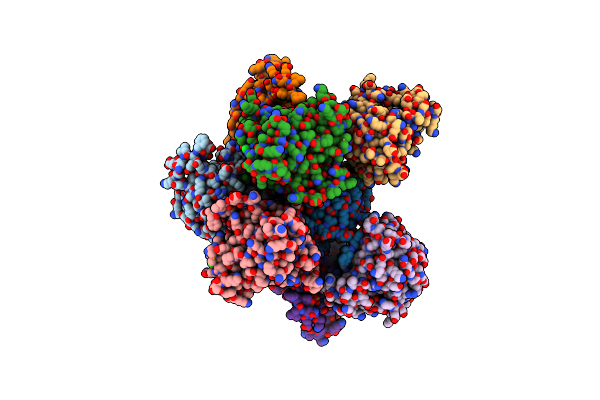 |
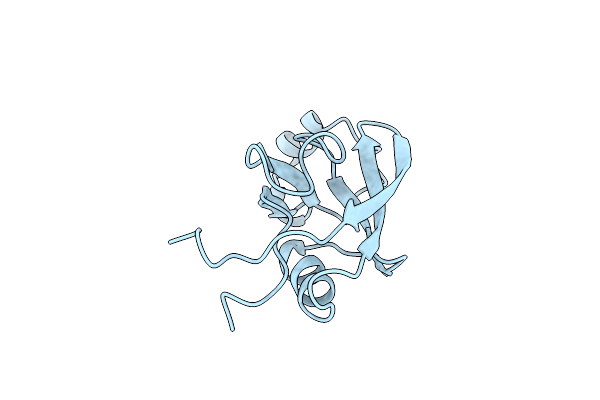
Crystal Structure Of Peptidyl-Prolyl Cis-Trans Isomerasefrom (Ppib) Streptococcus Pneumoniae R6
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2021-12-01 Classification: ISOMERASE Ligands: CL, EDO, MES |
 |
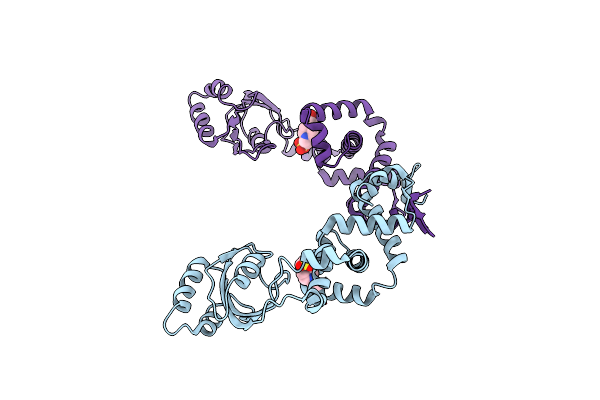
Crystal Structure Of The Pdz Domain Of The Serine Peptidase Htra From Streptococcus Agalactiae.
Organism: Streptococcus agalactiae a909
Method: X-RAY DIFFRACTION Resolution:0.97 Å Release Date: 2021-12-01 Classification: PROTEIN BINDING Ligands: CL |
 |
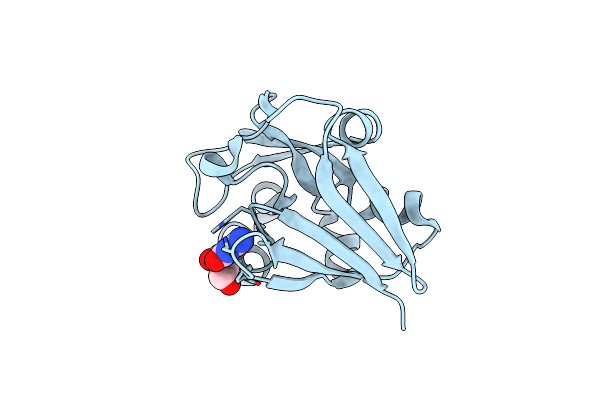
Crystal Structure Of Peptidylprolyl Isomerase Prsa From Streptococcus Mutans.
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2021-12-01 Classification: ISOMERASE Ligands: EPE, CL |
 |
High Resolution Crystal Structure Of Putative Pterin Binding Protein Prur (Vv2_1280) From Vibrio Vulnificus Cmcp6
Organism: Vibrio vulnificus (strain cmcp6)
Method: X-RAY DIFFRACTION Resolution:0.99 Å Release Date: 2021-11-17 Classification: Pterin binding protein Ligands: FMT, NA |
 |
High Resolution Crystal Structure Of Putative Pterin Binding Protein (Prur) From Vibrio Cholerae O1 Biovar El Tor Str. N16961 In Complex With Neopterin
Organism: Vibrio cholerae o1 biovar el tor str. n16961
Method: X-RAY DIFFRACTION Resolution:1.03 Å Release Date: 2021-11-17 Classification: PTERIN-BINDING PROTEIN Ligands: NEU |
 |
Crystal Structure Of The Peptidoglycan Binding Domain Of The Outer Membrane Protein (Ompa) From Klebsiella Pneumoniae With Bound D-Alanine
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2021-07-28 Classification: PEPTIDE BINDING PROTEIN Ligands: DAL, CL |

