Search Count: 28
 |
Crystal Structure Of The Shv-1 D104E Beta-Lactamase/Beta-Lactamase Inhibitor Protein (Blip) Complex
Organism: Klebsiella pneumoniae, Streptomyces clavuligerus
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2011-03-09 Classification: HYDROLASE/HYDROLASE INHIBITOR |
 |
Crystal Structure Of Tyrosine Aminotransferase Tripple Mutant (P181Q,R183G,A321K) From Escherichia Coli At 2.35 A Resolution
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2009-01-27 Classification: TRANSFERASE Ligands: PLR |
 |
Crystal Structure Of The Shv-1 Beta-Lactamase/Beta-Lactamase Inhibitor Protein (Blip) E73M/S130K/S146M Complex
Organism: Klebsiella pneumoniae, Streptomyces clavuligerus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-05-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: SO4 |
 |
Crystal Structure Of The Shv-1 Beta-Lactamase/Beta-Lactamase Inhibitor Protein (Blip) E73M Complex
Organism: Klebsiella pneumoniae, Streptomyces clavuligerus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2008-05-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: SO4 |
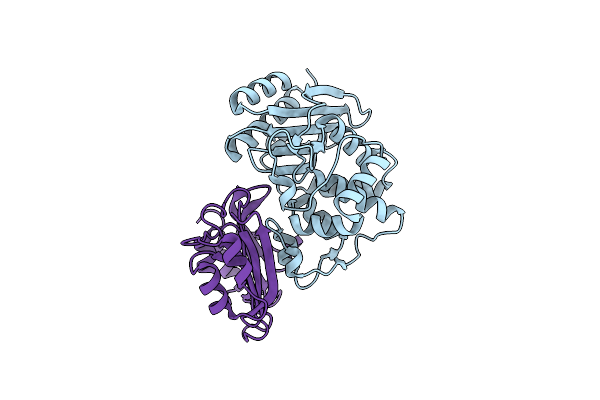 |
Crystal Structure Of The Shv-1 Beta-Lactamase/Beta-Lactamase Inhibitor Protein (Blip) Complex
Organism: Klebsiella pneumoniae, Streptomyces clavuligerus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2006-07-04 Classification: HYDROLASE/HYDROLASE INHIBITOR |
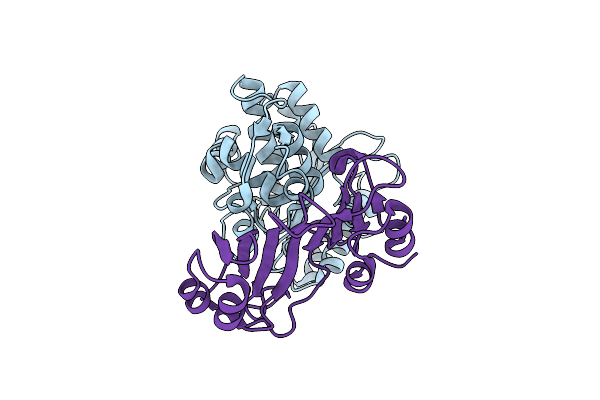 |
Crystal Structure Of The Shv D104K Beta-Lactamase/Beta-Lactamase Inhibitor Protein (Blip) Complex
Organism: Klebsiella pneumoniae, Streptomyces clavuligerus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-07-04 Classification: HYDROLASE/HYDROLASE INHIBITOR |
 |
Organism: Malus x domestica
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2005-05-03 Classification: LYASE Ligands: NI, K, PY4, TRS |
 |
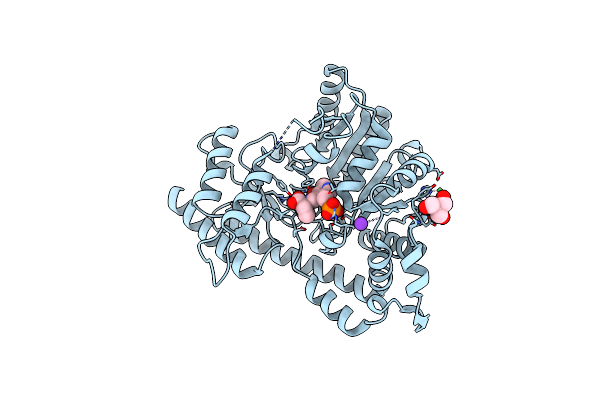
Unliganded Structure Of Hexamutant + A293D Mutant Of E. Coli Aspartate Aminotransferase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-10-05 Classification: TRANSFERASE Ligands: SO4 |
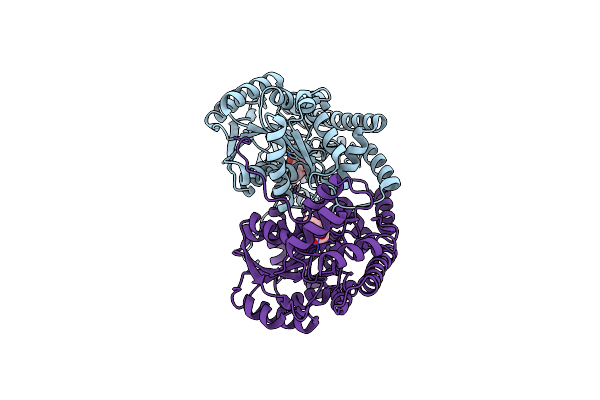 |
Hydrocinnamic Acid-Bound Structure Of Srhept + A293D Mutant Of E. Coli Aspartate Aminotransferase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2004-10-05 Classification: TRANSFERASE Ligands: HCI |
 |
Hydrocinnamic Acid-Bound Structure Of Hexamutant + A293D Mutant Of E. Coli Aspartate Aminotransferase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-10-05 Classification: TRANSFERASE Ligands: HCI |
 |
Hydrocinnamic Acid-Bound Structure Of Srhept Mutant Of E. Coli Aspartate Aminotransferase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-10-05 Classification: TRANSFERASE Ligands: HCI |
 |
Maleic Acid-Bound Structure Of Srhept Mutant Of E. Coli Aspartate Aminotransferase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2004-10-05 Classification: TRANSFERASE Ligands: MAE |
 |
Crystal Structure Of The R253K Mutant Of 7,8-Diaminopelargonic Acid Synthase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2004-03-23 Classification: TRANSFERASE |
 |
Crystal Structure Of The R253A Mutant Of 7,8-Diaminopelargonic Acid Synthase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2004-03-23 Classification: TRANSFERASE |
 |
Crystal Structure Of The D147N Mutant Of 7,8-Diaminopelargonic Acid Synthase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-03-23 Classification: TRANSFERASE |
 |
Crystal Structure Of The Y144F Mutant Of 7,8-Diaminopelargonic Acid Synthase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2004-03-23 Classification: TRANSFERASE |
 |
Crystal Structure Of The Y17F Mutant Of 7,8-Diaminopelargonic Acid Synthase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2004-03-23 Classification: TRANSFERASE |
 |
Crystal Structure Of Apple Acc Synthase In Complex With [2-(Amino-Oxy)Ethyl](5'-Deoxyadenosin-5'-Yl)(Methyl)Sulfonium
Organism: Malus x domestica
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2003-04-22 Classification: LYASE Ligands: PLP, AAD, MES |
 |
Crystal Structure Of Apple Acc Synthase In Complex With L-Aminoethoxyvinylglycine
Organism: Malus x domestica
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2002-12-23 Classification: LYASE Ligands: PPG, MRD |
 |
Crystal Structure Of The R391A Mutant Of 7,8-Diaminopelargonic Acid Synthase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2002-11-27 Classification: TRANSFERASE |

