Search Count: 37
 |
Solution Structure Of The Human Prostate Stem Cell Antigen (Psca), Water-Soluble Domain
|
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-04-02 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Alpha Sars-Cov-2 Spike Protein Rbd-Down In Complex With Regn10987 Fab Homologue (Local Refinement)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.60 Å Release Date: 2025-04-02 Classification: VIRAL PROTEIN |
 |
Serine Protease Inhibitor Hciq2C1 From Heteractis Crispa With Trpa1 Mediated Antinociceptive Activity
|
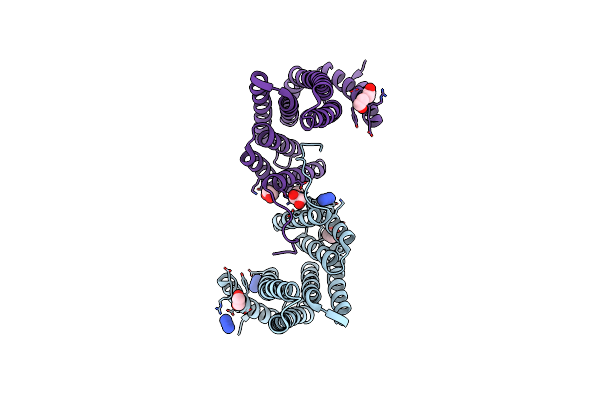 |
Crystal Structure Of The Chimera Of Human 14-3-3 Zeta And Phosphorylated Cytoplasmic Loop Fragment Of The Alpha7 Acetylcholine Receptor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-10-18 Classification: SIGNALING PROTEIN Ligands: EDO, AZI, BEZ, PEG |
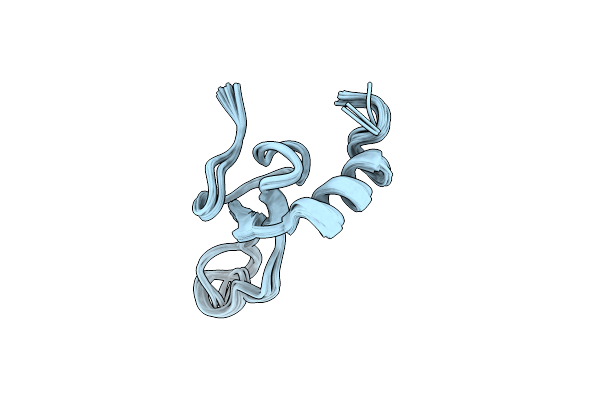 |
Spider Toxin Pha1B (Pntx3-6) From Phoneutria Nigriventer Targeting Cav2.X Calcium Channels And Trpa1 Channel
Organism: Phoneutria nigriventer
Method: SOLUTION NMR Release Date: 2023-06-21 Classification: TOXIN |
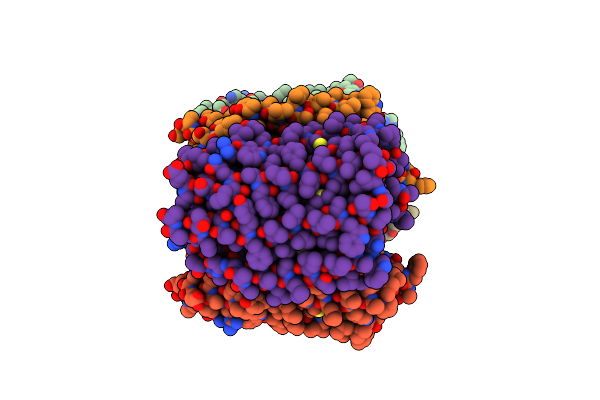 |
Crystal Structure Of The Microbial Rhodopsin From Sphingomonas Paucimobilis (Spar)
Organism: Sphingomonas paucimobilis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-05-03 Classification: MEMBRANE PROTEIN Ligands: LFA |
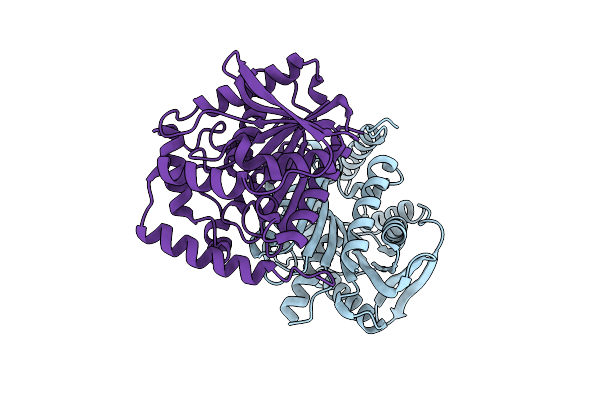 |
Crystal Structure Of Cold-Active Esterase Pmgl3 From Permafrost Metagenomic Library
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-11-03 Classification: HYDROLASE |
 |
 |
 |
Structure Of The Trans-(Tyr39-Pro40) Form Of The Human Secreted Ly-6/Upar Related Protein-1 (Slurp-1)
|
 |
Structure Of The Cis-(Tyr39-Pro40) Form Of The Human Secreted Ly-6/Upar Related Protein-1 (Slurp-1)
|
 |
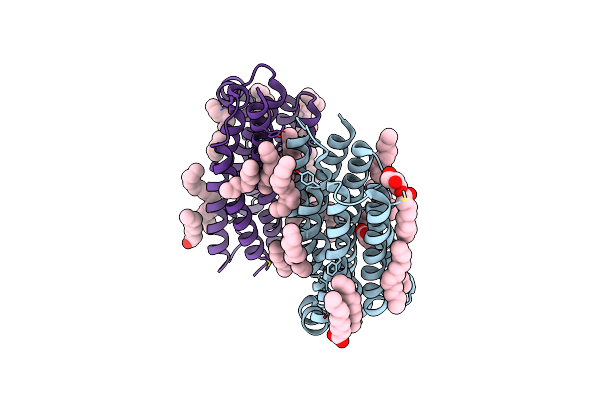
Organism: Organic lake phycodnavirus
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2020-11-25 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
Organism: Organic lake phycodnavirus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-11-25 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, OLC |
 |
Organism: Organic lake phycodnavirus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-11-25 Classification: MEMBRANE PROTEIN Ligands: LFA, 97N |
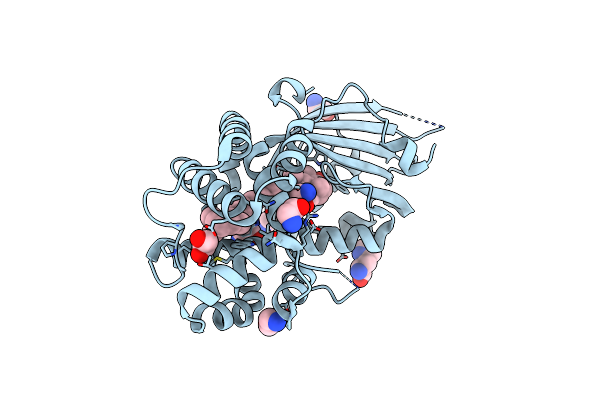 |
Organism: Synechocystis sp. (strain pcc 6803 / kazusa)
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2020-11-18 Classification: PLANT PROTEIN Ligands: ECH, GLY, CL, IMD, HIS, EDO |
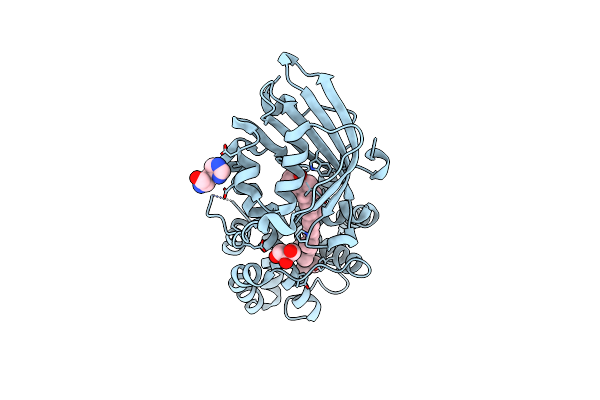 |
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2020-11-18 Classification: PLANT PROTEIN Ligands: ECH, GOL, HIS |
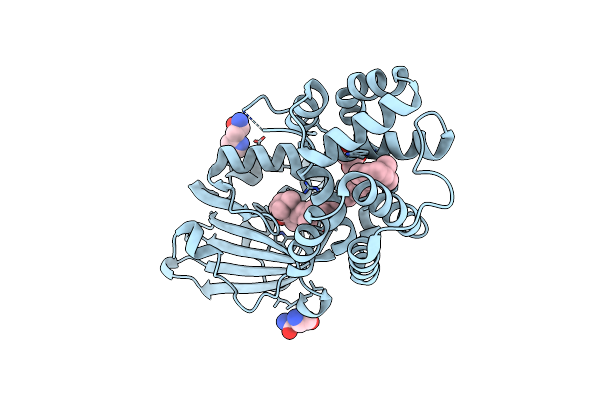 |
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-11-18 Classification: PLANT PROTEIN Ligands: ASN, ECH, CL, GOL, HIS |
 |
Crystal Structure Of C173S Mutation In The Pmgl2 Esterase From Permafrost Metagenomic Library
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-07-08 Classification: HYDROLASE Ligands: MG, PG6 |
 |
Crystal Structure Of The N112A Mutant Of The Light-Driven Sodium Pump Kr2 In The Pentameric Form, Ph 8.0
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-06-17 Classification: MEMBRANE PROTEIN Ligands: OLC, OLA, LFA, BOG, RET, NA |

