Search Count: 16
 |
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2012-09-19 Classification: TRANSFERASE Ligands: CIT |
 |
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-09-19 Classification: TRANSFERASE |
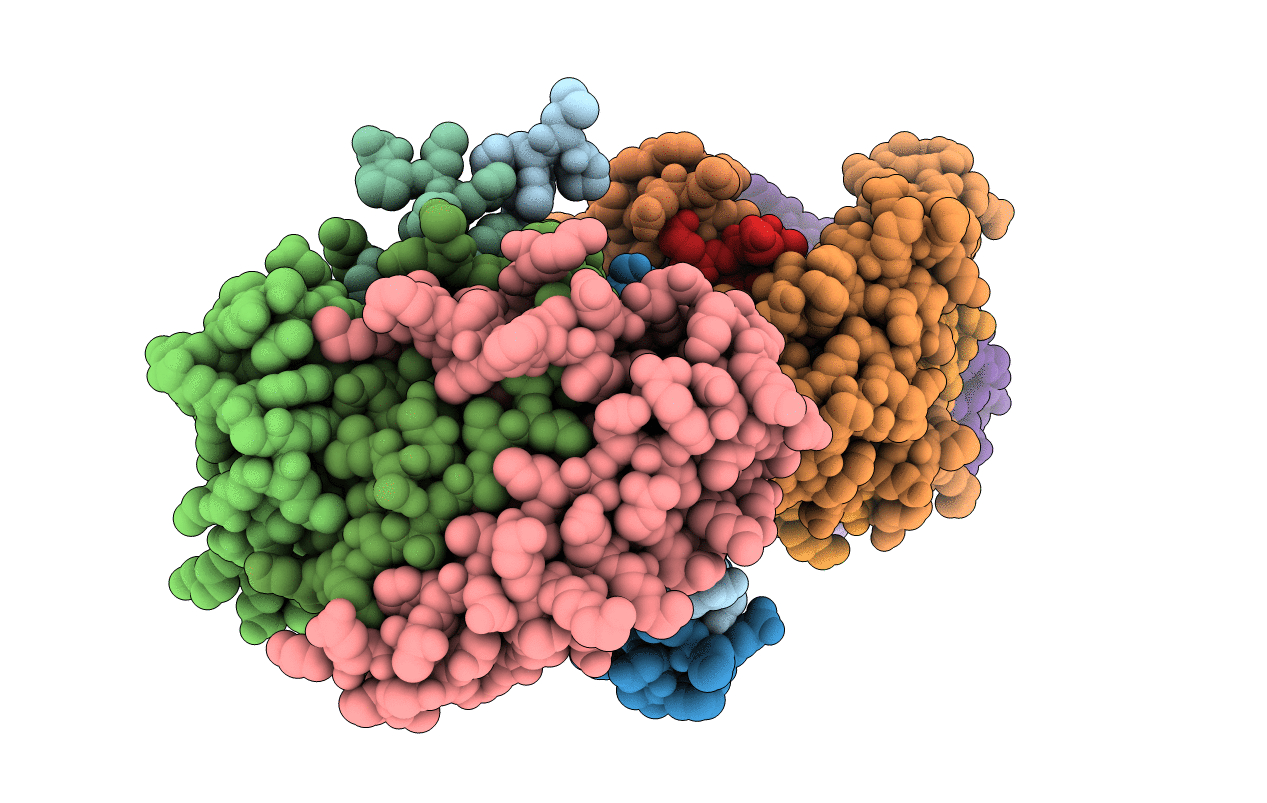 |
E109Q Mutant Of Pyruvoyl-Dependent Arginine Decarboxylase From Methanococcus Jannashii
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-03-18 Classification: LYASE Ligands: AG2, MPD |
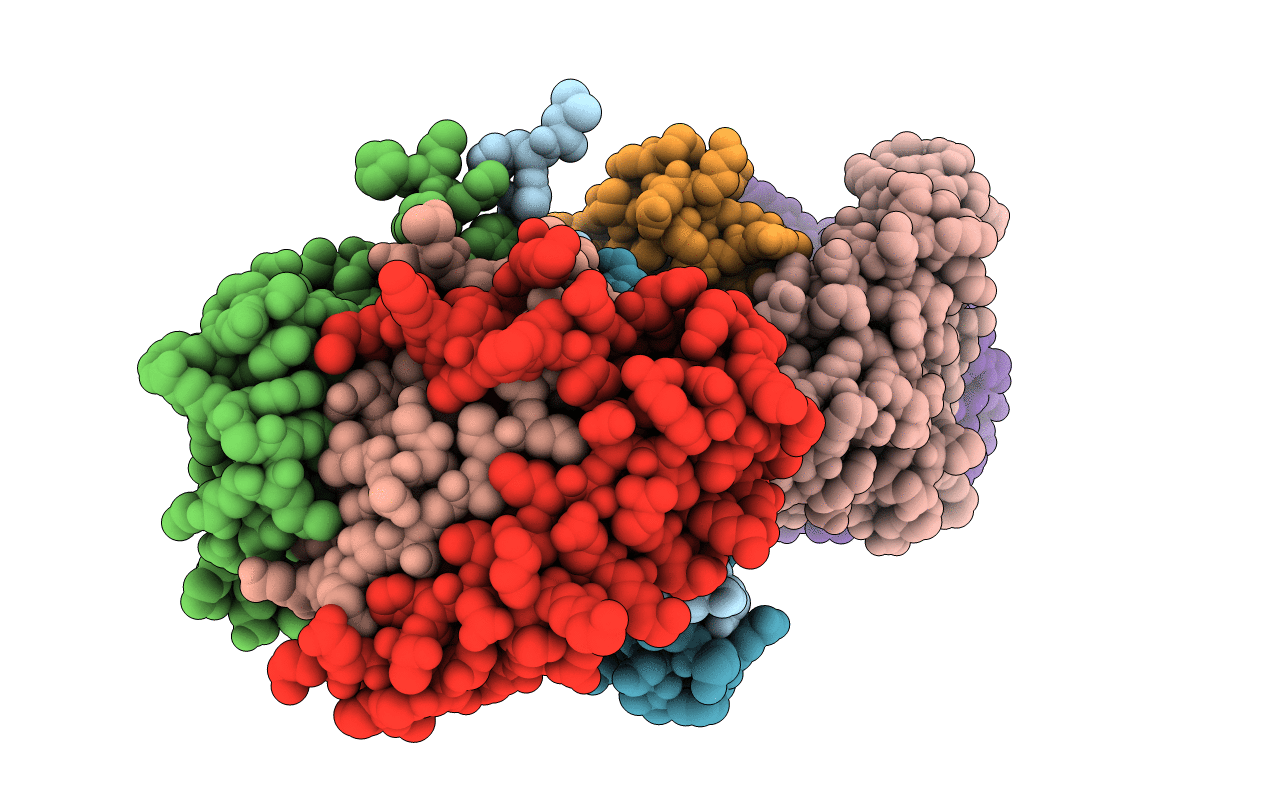 |
N47A Mutant Of Pyruvoyl-Dependent Arginine Decarboxylase From Methanococcus Jannashii
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-03-18 Classification: LYASE Ligands: AG2, PYR, MPD |
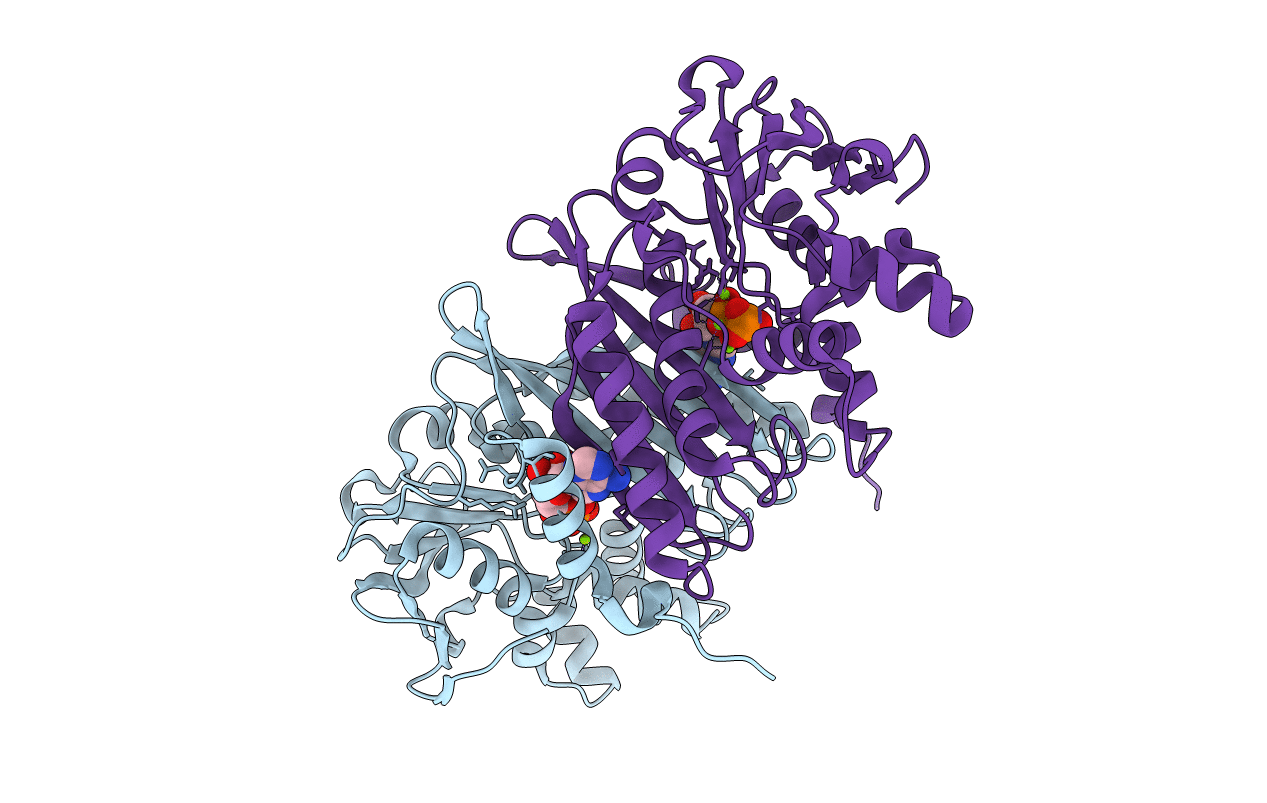 |
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-03-18 Classification: TRANSFERASE Ligands: MG, ATP |
 |
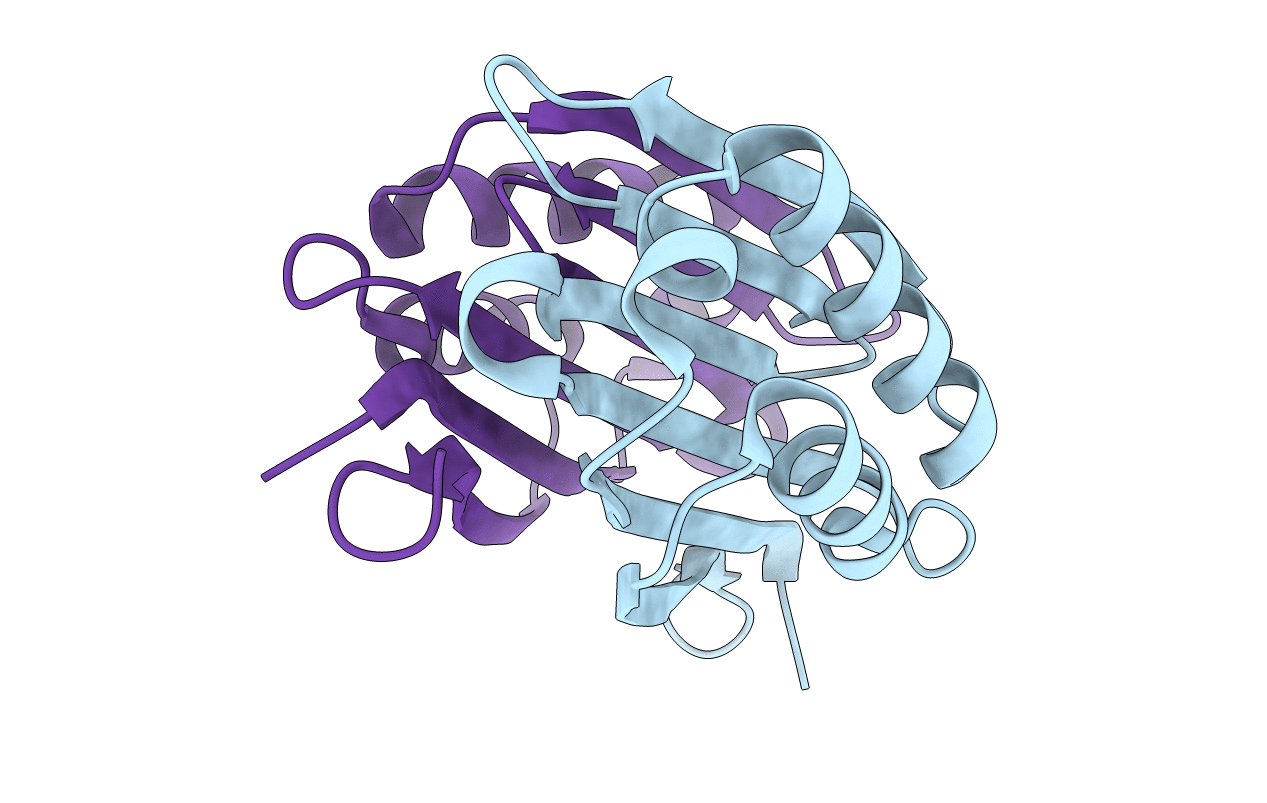
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-03-18 Classification: TRANSFERASE Ligands: MG, ACP |
 |
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2008-03-18 Classification: TRANSFERASE Ligands: MG, TPS, ACP |
 |
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2008-03-18 Classification: TRANSFERASE Ligands: MG, TPP, ADP |
 |
Crystal Structure Of Thermotoga Maritima S-Adenosylmethionine Decarboxylase
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2004-06-29 Classification: LYASE |
 |
Structure Of Thermotoga Maritima S63A Non-Processing Mutant S-Adenosylmethionine Decarboxylase
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2004-06-29 Classification: LYASE |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2002-07-03 Classification: METAL BINDING PROTEIN Ligands: MN, MG, FMT |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2002-07-03 Classification: METAL BINDING PROTEIN Ligands: MN, MG, FMT |
 |
Crystal Structure Of Orotidine 5'-Monophosphate Decarboxylase From Bacillus Subtilis Complexed With Ump
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2000-03-06 Classification: LYASE Ligands: U5P |
 |
Organism: Paenibacillus thiaminolyticus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1999-10-07 Classification: TRANSFERASE Ligands: SO4 |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1998-10-14 Classification: TRANSFERASE Ligands: SO4 |
 |
Thiaminase I From Bacillus Thiaminolyticus With Covalently Bound 4-Amino-2,5-Dimethylpyrimidine
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1998-10-14 Classification: TRANSFERASE Ligands: SO4, PYD |

